Search Count: 2,339
All
Selected
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2026-02-18 Classification: HYDROLASE Ligands: ZN, MG, A1EL5, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2026-02-18 Classification: HYDROLASE Ligands: ZN, MG, A1E46 |
 |
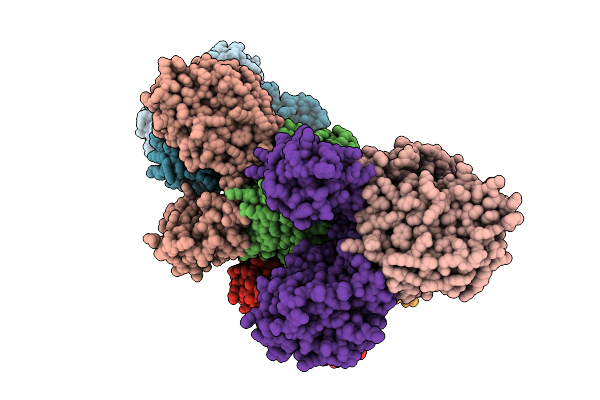
Organism: Homo sapiens, Bos taurus
Method: ELECTRON MICROSCOPY Resolution:4.40 Å Release Date: 2026-02-18 Classification: SIGNALING PROTEIN Ligands: ME7, CA |
 |
Organism: Homo sapiens, Bos taurus
Method: ELECTRON MICROSCOPY Resolution:7.00 Å Release Date: 2026-02-18 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens, Bos taurus
Method: ELECTRON MICROSCOPY Resolution:4.50 Å Release Date: 2026-02-18 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2026-02-11 Classification: HYDROLASE Ligands: ZN, MG, A1EYL |
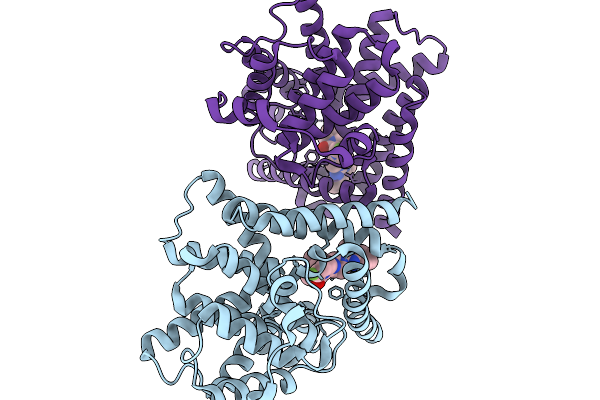 |
Crystal Structure Of Human Phosphodiesterase 10A In Complex With (6-Fluoro-2-(1-(4-Methylquinazolin-2-Yl)Azetidin-3-Yl)Imidazo[1,2-A]Pyridin-3-Yl)(4,7-Diazaspiro[2.5]Octan-7-Yl)Methanone
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: A1ETB, MG, ZN |
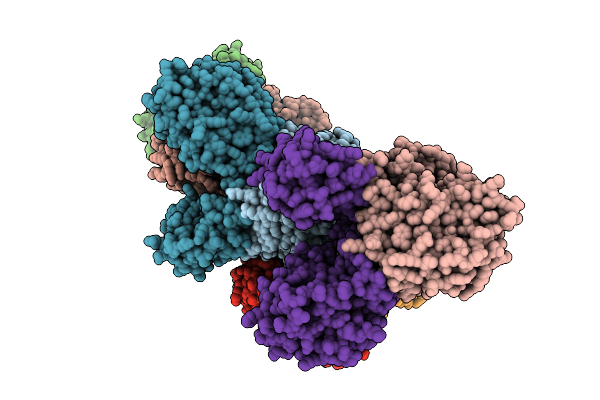 |
Organism: Borreliella burgdorferi
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2026-01-21 Classification: HYDROLASE Ligands: MN, FE, AMP |
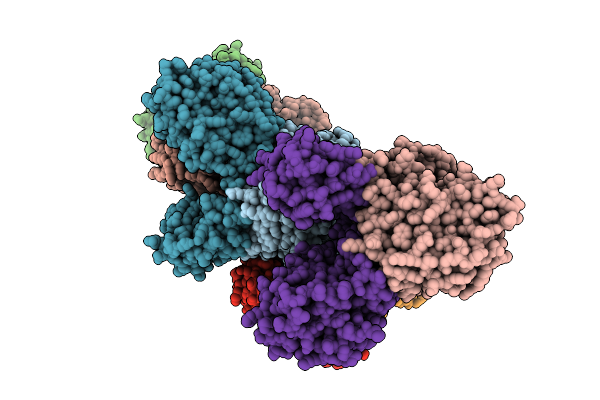 |
Organism: Borreliella burgdorferi
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2026-01-21 Classification: HYDROLASE Ligands: MN, FE, AMP |
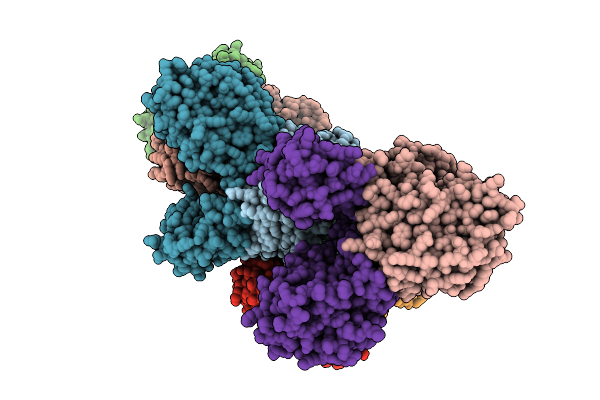 |
Organism: Borreliella burgdorferi
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2026-01-21 Classification: HYDROLASE Ligands: MN, FE, AMP |
 |
Organism: Borreliella burgdorferi
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2026-01-21 Classification: HYDROLASE Ligands: MN, FE, A1JDM |
 |
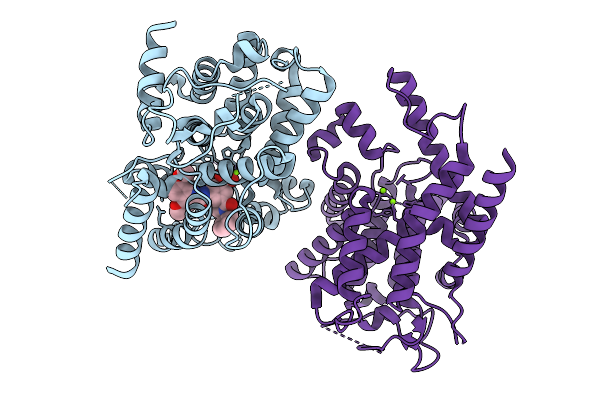
Human Ectonucleotide Pyrophosphatase/Phosphodiesterase Family Member 3 (Enpp3) Inhibitor Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2026-01-07 Classification: HYDROLASE Ligands: A1BYU, ZN, CA, NAG, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-12-31 Classification: HYDROLASE Ligands: X5W, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2025-12-24 Classification: LIPID BINDING PROTEIN Ligands: A1IWC, GOL, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-12-24 Classification: LIPID BINDING PROTEIN Ligands: A1IWD |
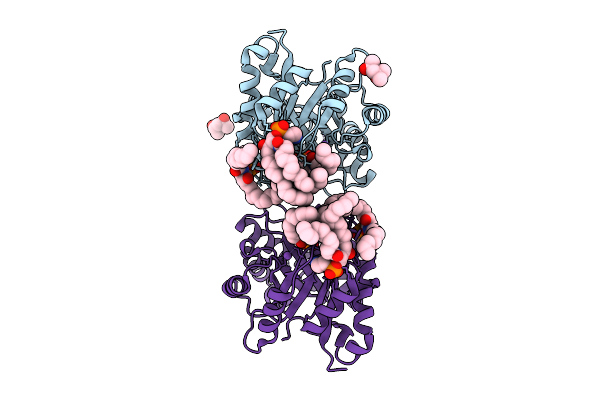 |
Structure Of Phospholipase D Betaib1I From Sicarius Terrosus Venom, H47N Mutant Bound To Product And Substrate Sphingolipids At 2.2 A Resolution From A 2-Day Old Crystal
Organism: Sicarius terrosus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-12-03 Classification: LYASE Ligands: A1A43, A1A44, NA, MG, MPD |
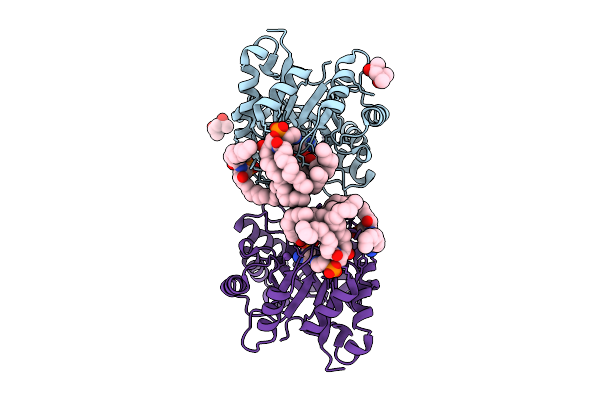 |
Structure Of Phospholipase D Betaib1I From Sicarius Terrosus Venom, H47N Mutant Bound To Substrate Sphingolipids At 2.60 A Resolution
Organism: Sicarius terrosus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-12-03 Classification: LYASE Ligands: A1A43, NA, MG, MPD |
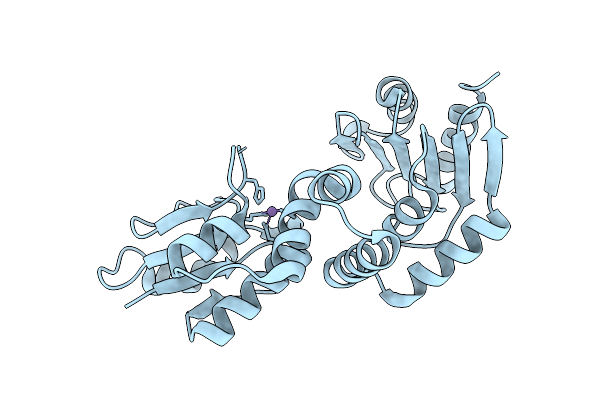 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: MN |
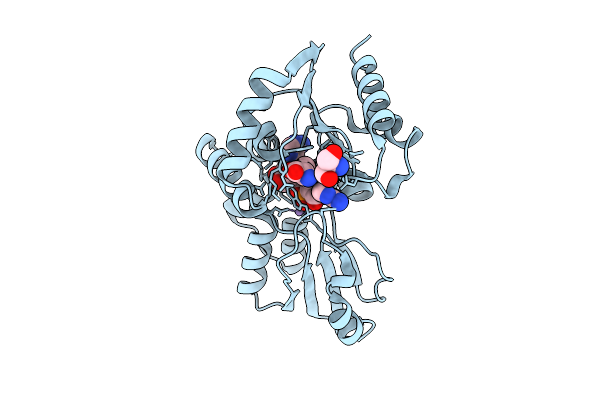 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: A1EL3, TRS, MN |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: A1EL0, MN |

