Search Count: 7,928
All
Selected
 |
Pathogen Effector Forms A Phosphatase Holoenzyme Complex With Host Core Enzyme To Promote Disease
Organism: Phytophthora sojae, Homo sapiens, Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2026-03-04 Classification: PLANT PROTEIN Ligands: MN |
 |
Pathogen Effector Forms A Phosphatase Holoenzyme Complex With Host Core Enzyme To Promote Disease
Organism: Homo sapiens, Phytophthora sojae, Arabidopsis thaliana, Peptidiphaga
Method: ELECTRON MICROSCOPY Release Date: 2026-03-04 Classification: PLANT PROTEIN Ligands: MN |
 |
Pathogen Effector Forms A Phosphatase Holoenzyme Complex With Host Core Enzyme To Promote Disease
Organism: Phytophthora sojae, Arabidopsis thaliana, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-03-04 Classification: PLANT PROTEIN Ligands: MN |
 |
Pathogen Effector Forms A Phosphatase Holoenzyme Complex With Host Core Enzyme To Promote Disease
Organism: Arabidopsis thaliana, Phytophthora sojae, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-03-04 Classification: PLANT PROTEIN Ligands: MN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: A1IIY, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2026-01-28 Classification: HYDROLASE |
 |
Crystal Structure Of Oxidized Form (C92-C121) Of Protein Tyrosine Phosphatase 1B (Ptp1B)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2026-01-28 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2026-01-28 Classification: HYDROLASE |
 |
Structure Of Human Wild-Type Signal Regulatory Protein Alpha V2 Variant (Sirpav2) With Zinc
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2026-01-28 Classification: IMMUNE SYSTEM Ligands: ZN, SO4 |
 |
Cryo-Em Structure Of Human Lipid Phosphate Phosphatase 1 Complexed With Po4 In Nanodiscs
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.82 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN Ligands: LPP, NAG, LBN, PT5, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2026-01-28 Classification: IMMUNE SYSTEM Ligands: EOH, GOL, SO4, A1CU3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2026-01-28 Classification: IMMUNE SYSTEM Ligands: GOL, EOH, MG, A1CWM, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2026-01-28 Classification: IMMUNE SYSTEM Ligands: EOH, A1CWL, SO4, GOL, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2026-01-28 Classification: IMMUNE SYSTEM Ligands: CL, EDO, GOL, A1CWK, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2026-01-28 Classification: IMMUNE SYSTEM Ligands: EDO, NA, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2026-01-21 Classification: TRANSFERASE Ligands: MN |
 |
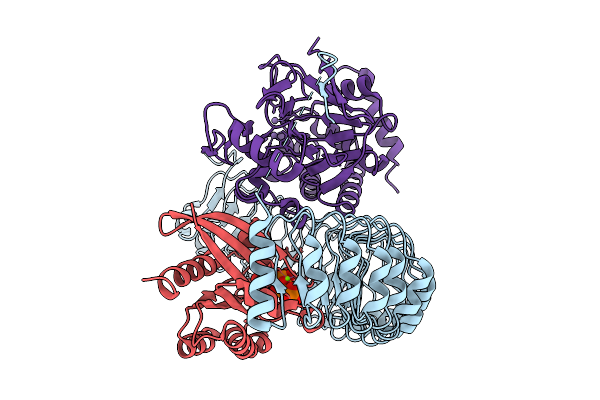
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.50 Å Release Date: 2026-01-21 Classification: STRUCTURAL PROTEIN/Hydrolase Ligands: MN |
 |
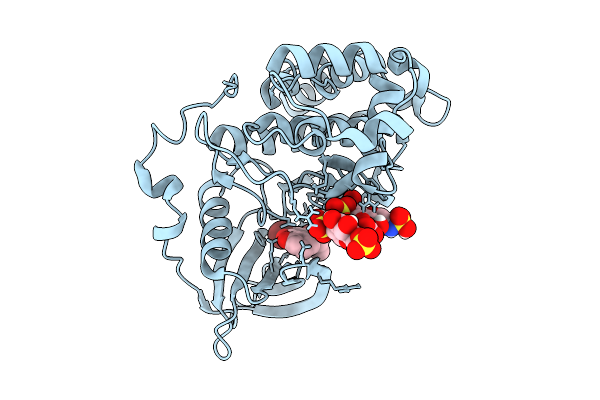
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2026-01-21 Classification: SIGNALING PROTEIN Ligands: GNP, MG, MN |
 |
Ck2, Catalytic Subunit Alpha' (Csnk2A2 Gene Product) In Complex With A Heparin-Derived Trisaccharide And The Atp-Competitive Inhibitor 4W
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2026-01-21 Classification: TRANSFERASE Ligands: A1I5N, CL |
 |
Structure Of Rv2985, A Nudix Diadenosine Polyphosphatase From M. Tuberculosis, In Complex With Putative Atp
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2026-01-14 Classification: CELL INVASION Ligands: ATP, SO4 |

