Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Bacillus phage phi3t
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2024-07-24 Classification: SIGNALING PROTEIN Ligands: A1H22, FU1 |
 |
Organism: Bacillus phage phi3t
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-07-24 Classification: SIGNALING PROTEIN Ligands: A1H22 |
 |
Organism: Bacillus phage phi3t
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2024-07-24 Classification: SIGNALING PROTEIN Ligands: A1H22 |
 |
Organism: Bacillus phage phi3t
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2024-07-24 Classification: SIGNALING PROTEIN Ligands: A1H22 |
 |
Organism: Ostrinia furnacalis
Method: SOLUTION NMR Release Date: 2023-02-15 Classification: SIGNALING PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-03-16 Classification: MEMBRANE PROTEIN Ligands: NAG, Y01 |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-03-16 Classification: MEMBRANE PROTEIN Ligands: NAG, Y01 |
 |
Structure Of The Gpcr Dimer Ste2 In The Inactive-Like State Bound To Agonist
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-03-16 Classification: MEMBRANE PROTEIN Ligands: NAG, Y01 |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-03-16 Classification: MEMBRANE PROTEIN Ligands: NAG, Y01 |
 |
Organism: Helicoverpa armigera
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-02-02 Classification: TRANSPORT PROTEIN |
 |
Organism: Helicoverpa armigera
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-02-02 Classification: TRANSPORT PROTEIN |
 |
Organism: Helicoverpa armigera
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-02-02 Classification: TRANSPORT PROTEIN Ligands: 81K |
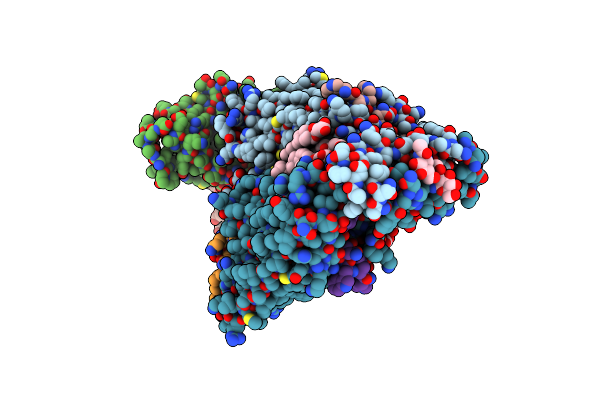 |
Organism: Saccharomyces cerevisiae, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: MEMBRANE PROTEIN Ligands: NAG, Y01 |
 |
Cryo-Em Structure Of The Signal Sequence-Engaged Post-Translational Sec Translocon
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2020-12-02 Classification: MEMBRANE PROTEIN |
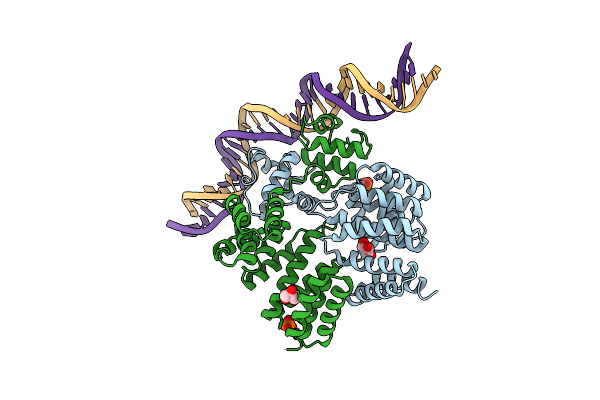 |
Crystal Structure Of Streptococcus Dysgalactiae Shp Pheromone Receptor Rgg2 Bound To Dna
Organism: Streptococcus dysgalactiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-10-21 Classification: DNA BINDING PROTEIN/DNA Ligands: PO4, GOL |
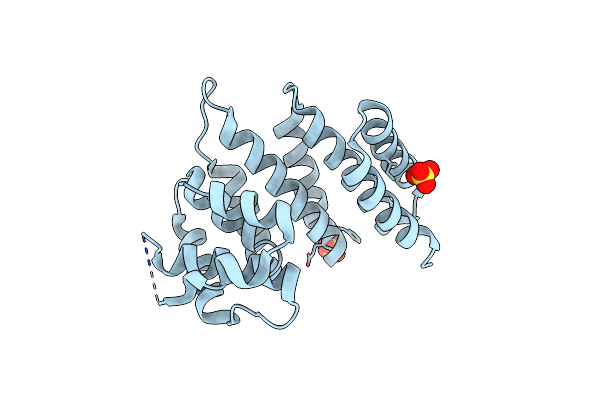 |
Crystal Structure Of Streptococcus Thermophilus Shp Pheromone Receptor Rgg3
Organism: Streptococcus thermophilus (strain atcc baa-250 / lmg 18311)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-10-21 Classification: DNA BINDING PROTEIN Ligands: SO4 |
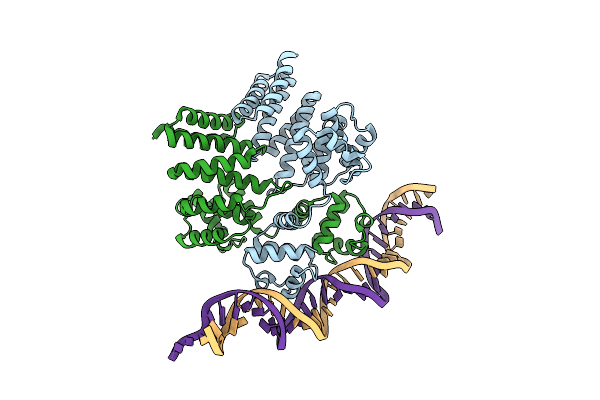 |
Crystal Structure Of Streptococcus Thermophilus Shp Pheromone Receptor Rgg3 Bound To Dna
Organism: Streptococcus thermophilus (strain atcc baa-250 / lmg 18311), Streptococcus thermophilus cnrz1066
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2020-10-21 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Epiphyas postvittana
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-10-14 Classification: TRANSPORT PROTEIN Ligands: PG0 |
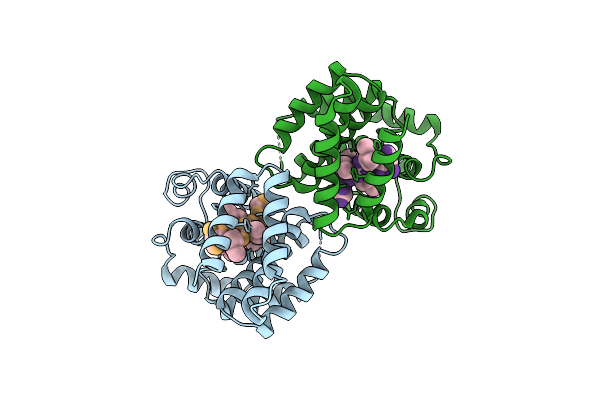 |
Cryoem Structure Of Streptococcus Thermophilus Shp Pheromone Receptor Rgg3 In Complex With Shp3
Organism: Streptococcus thermophilus (strain atcc baa-250 / lmg 18311), Streptococcus thermophilus cnrz1066
Method: ELECTRON MICROSCOPY Release Date: 2020-10-07 Classification: DNA BINDING PROTEIN |
 |
Organism: Lymantria dispar
Method: SOLUTION NMR Release Date: 2020-09-23 Classification: LIPID BINDING PROTEIN |