Search Count: 182
 |
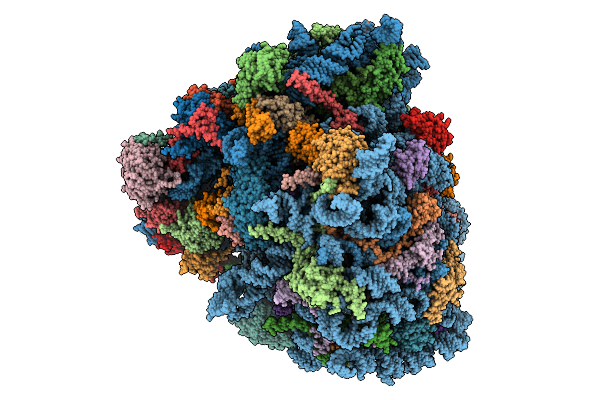
Gtpbp1*Gcp*Phe-Trna*Ribosome In The Gtpase Activation-Like State, Structure Iii
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, K, GCP, MG |
 |
Gtpbp1*Gdp*Phe-Trna*Ribosome In The Post-Gtp Hydrolysis State, Structure Iv
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, K, GDP, MG |
 |
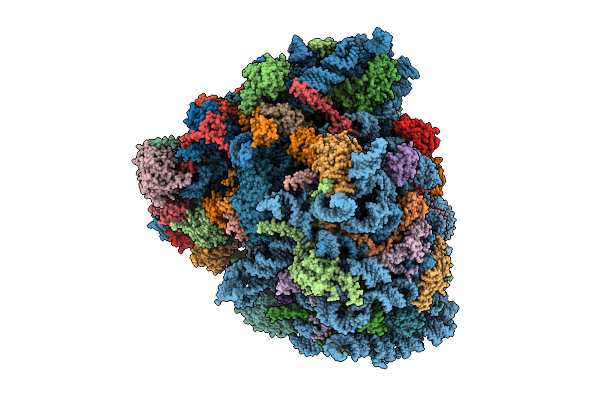
Organism: Oryctolagus cuniculus, Saccharomyces cerevisiae, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET |
 |
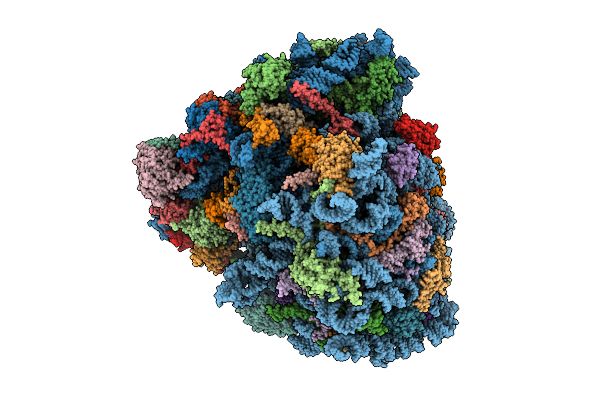
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, GCP, MG, K |
 |
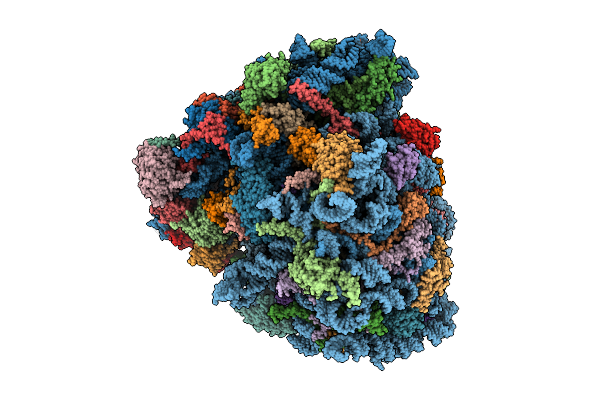
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, K, GCP, MG |
 |
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, GCP, MG, K |
 |
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, K, GCP, MG |
 |
Organism: Saccharomyces cerevisiae, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: PHE, GLY |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: BLOOD CLOTTING Ligands: CA, NA, CL, IMD, 0GJ, GLU, ILE, PHE, PTR |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: RIBOSOME Ligands: PHE, GNP, MG, ZN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2025-04-16 Classification: VIRAL PROTEIN, Hydrolase Ligands: GLY, VAL, THR, PHE, A1ALW |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:4.12 Å Release Date: 2025-02-12 Classification: TRANSPORT PROTEIN Ligands: PHE |
 |
Crystal Structure Of 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase (Dahp Synthase) From Providencia Alcalifaciens Complexed With Phe
Organism: Providencia alcalifaciens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-01-22 Classification: TRANSFERASE Ligands: PEG, PHE |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2025-01-01 Classification: OXIDOREDUCTASE Ligands: CO, PHE |
 |
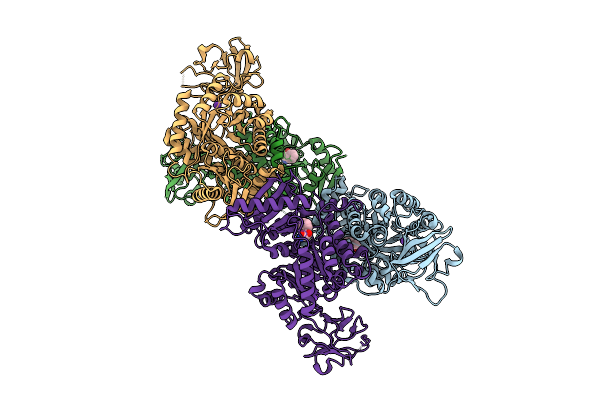
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-12-11 Classification: PROTEIN BINDING Ligands: PHE, ALA, NI, MG |
 |
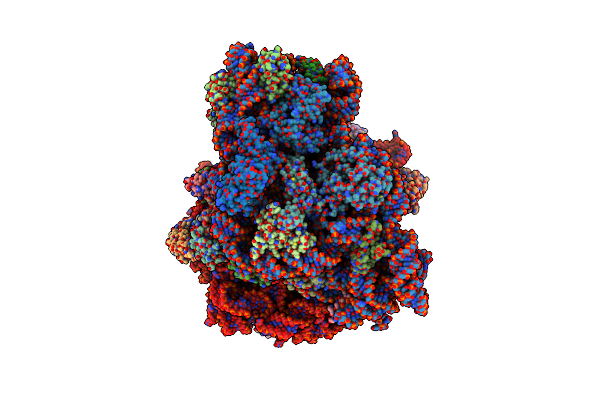
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2024-11-20 Classification: TRANSFERASE Ligands: K, PHE |
 |
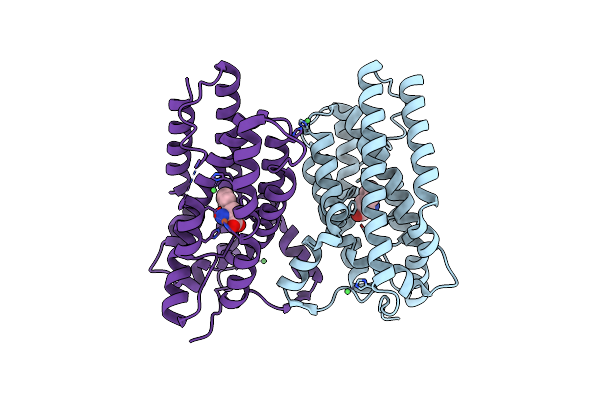
Organism: Escherichia coli, Synthetic construct, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: GENE REGULATION Ligands: ZN, K, MG, PHE |
 |
Organism: Aetokthonos hydrillicola thurmond2011
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2024-10-02 Classification: METAL BINDING PROTEIN Ligands: FE2, PHE, NI |
 |
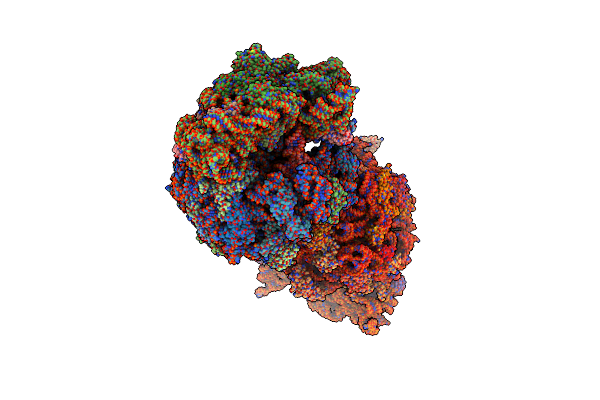
L-Amino Acid Oxidase 4 (Hclaao4) From The Fungus Hebeloma Cylindrosporum In Complex With L-Phenylalanine
Organism: Hebeloma cylindrosporum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-08-14 Classification: OXIDOREDUCTASE Ligands: FDA, PHE, SO4, PGR |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Macrolone Mcx-66, Mrna, Aminoacylated A-Site Phe-Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.40A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-08-07 Classification: RIBOSOME Ligands: MG, K, A1AEZ, ZN, SF4, PHE, FME |

