Search Count: 12
 |
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: LYASE Ligands: PGU |
 |
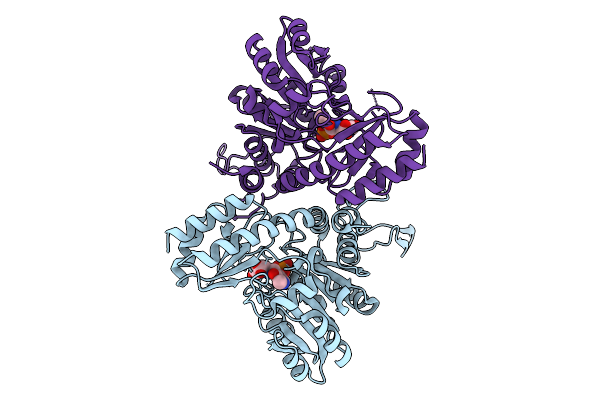
Crystal Structure Of A Heterooligomeric Aminotransferase From Serratia Sp. Atcc 39006, Ppe-Bound Form
Organism: Serratia sp. atcc 39006
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: PGU |
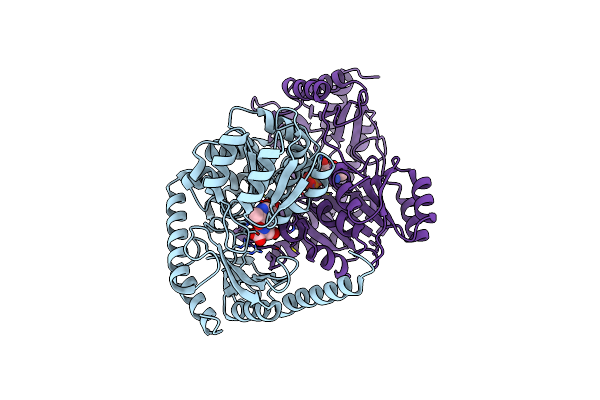 |
Crystal Structure Of Alpha-Oxoamine Synthase Alb29 With Plp Cofactor And L-Glutamate
Organism: Streptomyces albogriseolus 1-36
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2024-05-01 Classification: TRANSFERASE Ligands: PGU, PLP |
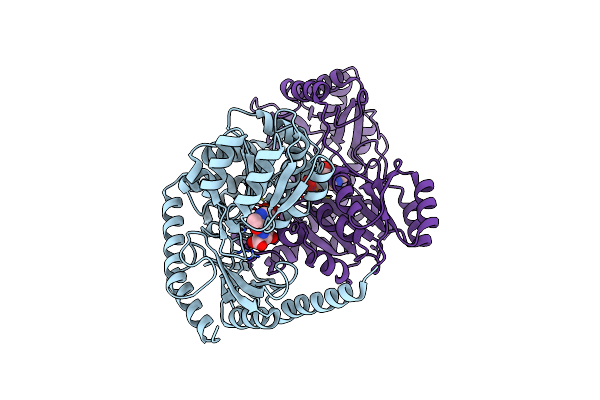 |
Crystal Structure Of Alpha-Oxoamine Synthase Alb29 With Plp Cofactor And L-Glutamate
Organism: Streptomyces albogriseolus 1-36
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-05-01 Classification: TRANSFERASE Ligands: PGU, PLP |
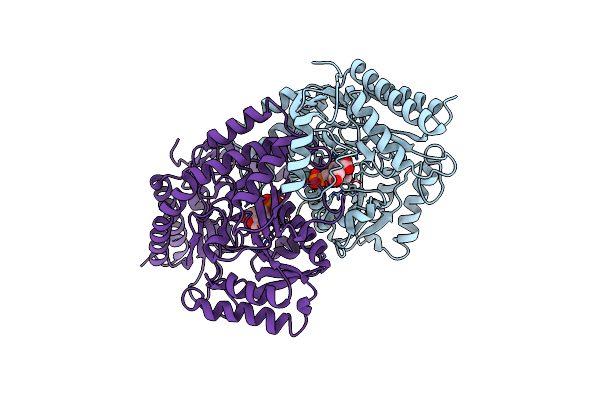 |
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2022-03-30 Classification: TRANSFERASE Ligands: PGU |
 |
Crystal Structure Of Lysn, Alpha-Aminoadipate Aminotransferase (Complexed With N-(5'-Phosphopyridoxyl)-L-Glutamate), From Thermus Thermophilus Hb27
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2009-09-01 Classification: TRANSFERASE Ligands: PGU |
 |
Crystal Structure Of Ll-Diaminopimelate Aminotransferase From Arabidopsis Thaliana Complexed With Plp-Glu: An External Aldimine Mimic
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2008-10-14 Classification: TRANSFERASE Ligands: PGU, GOL, SO4 |
 |
Crystal Structure Of Gdp-4-Keto-6-Deoxymannose-3-Dehydratase With A Trapped Plp-Glutamate Geminal Diamine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-12-18 Classification: TRANSFERASE Ligands: PGU |
 |
X-Ray Crystal Structure Of S. Venezuelae Desv In Complex With Ketimine Intermediate
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2007-05-15 Classification: HYDROLASE Ligands: CL, NA, EDO, PGU |
 |
Crystal Structure Of E.Coli Aspat Complexed With N-Phosphopyridoxyl-L-Glutamic Acid
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-06-14 Classification: TRANSFERASE Ligands: PGU |
 |
Crystal Structure Of Eschelichia Coli Branched-Chain Amino Acid Aminotransferase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2003-05-06 Classification: TRANSFERASE Ligands: PGU |
 |
Crystal Structures Of True Enzymatic Reaction Intermediates: Aspartate And Glutamate Ketimines In Aspartate Aminotransferase
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1994-01-31 Classification: AMINOTRANSFERASE Ligands: PGU |

