Search Count: 62
 |
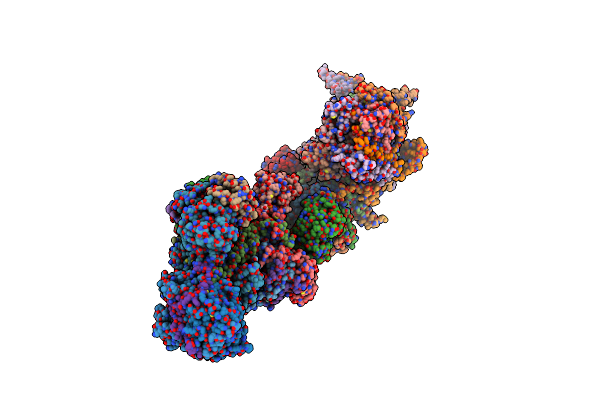
Organism: Pseudomonas aeruginosa pao1
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: J4U, PGT, PTY |
 |
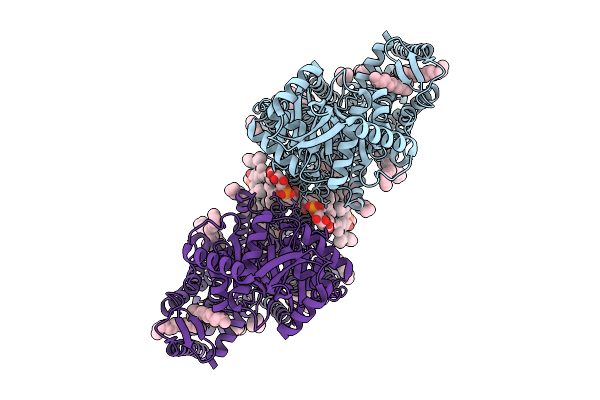
Organism: Pseudomonas aeruginosa pao1
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: J4U, PGT, PTY |
 |
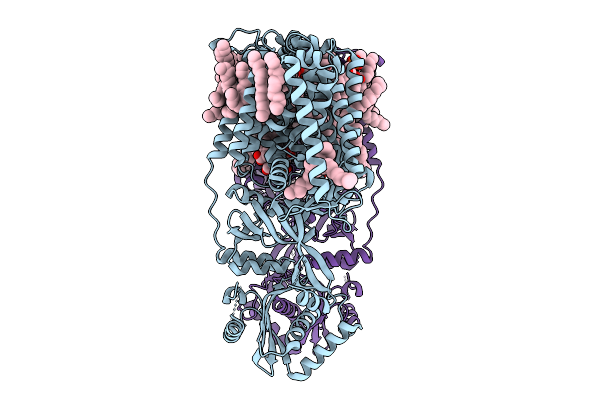
Organism: Pseudomonas aeruginosa pao1
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: PGT |
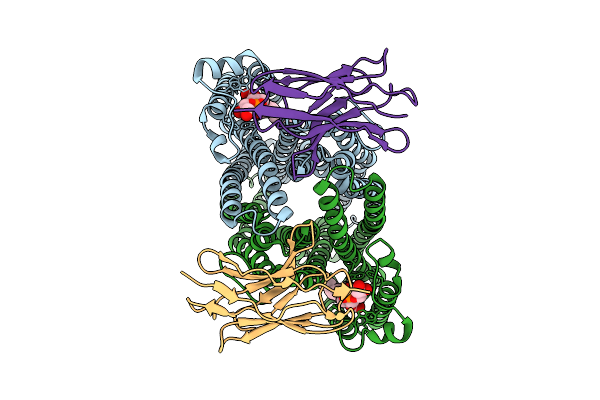 |
Cryo-Em Structure Of Sbma In The Inward-Facing-Narrow Conformation Bound To 2 Sybodies
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: TRANSPORT PROTEIN Ligands: PGT |
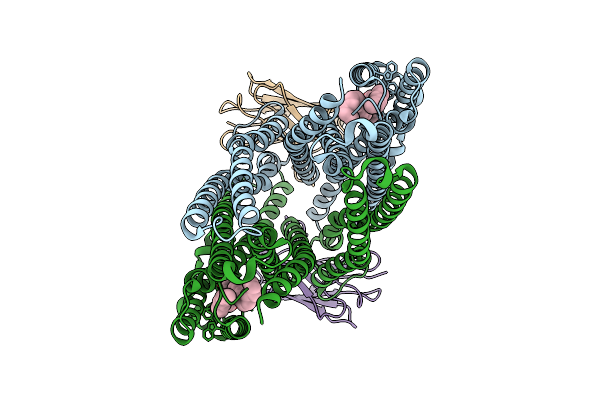 |
Cryo-Em Structure Of Sbma In The Inward-Facing-Wide Conformation Bound To 2 Sybodies
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: TRANSPORT PROTEIN Ligands: PGT |
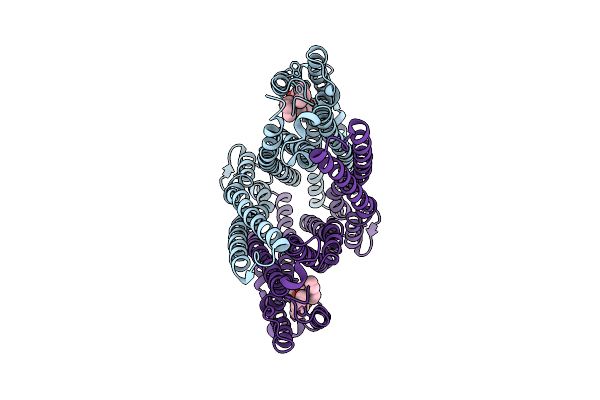 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: TRANSPORT PROTEIN Ligands: PGT |
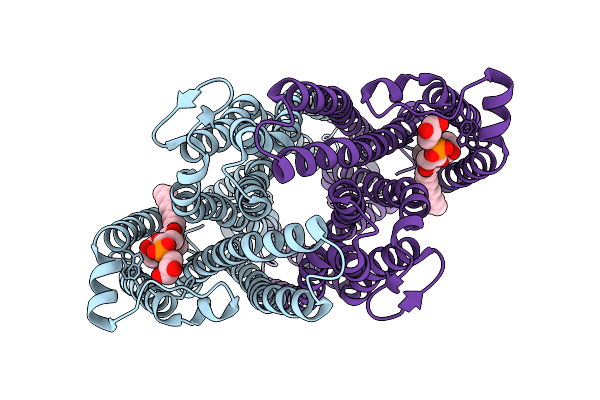 |
Cryo-Em Structure Of Sbma In The Inward-Facing-Occluded Conformation In Lipid Nanodiscs
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: TRANSPORT PROTEIN Ligands: PGT |
 |
Structure Of The Chlamydomonas Reinhardtii Respiratory Supercomplex I1 Iii2 Iv2
|
 |
Structure Of The Chlamydomonas Reinhardtii Respiratory Complex I From Respiratory Supercomplex
Organism: Chlamydomonas reinhardtii
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: FES, FMN, SF4, PTY, 8Q1, NDP, UQ5, PC7, 3PH, CDL, PGT, COO, ZN |
 |
Structure Of The Chlamydomonas Reinhardtii Respiratory Complex Iv From Respiratory Supercomplex
Organism: Chlamydomonas reinhardtii
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: HEA, CU, MG, PC7, PGT, CUA, PTY, 3PH |
 |
Structure Of The Chlamydomonas Reinhardtii Respiratory Complex Iv From Respiratory Supercomplex
Organism: Chlamydomonas reinhardtii
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: HEA, CU, MG, PC7, PGT, CUA, PTY, 3PH |
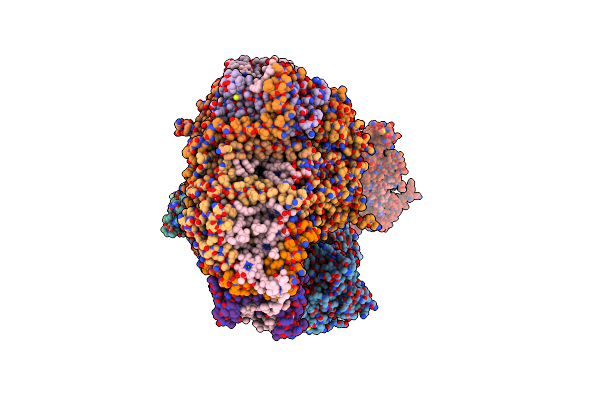 |
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: ELECTRON TRANSPORT Ligands: BCR, PGT, A1H1M, LMG, PQ9, SQD, SF4, CLA, DGD, CL0, PQN, CHL, LUT |
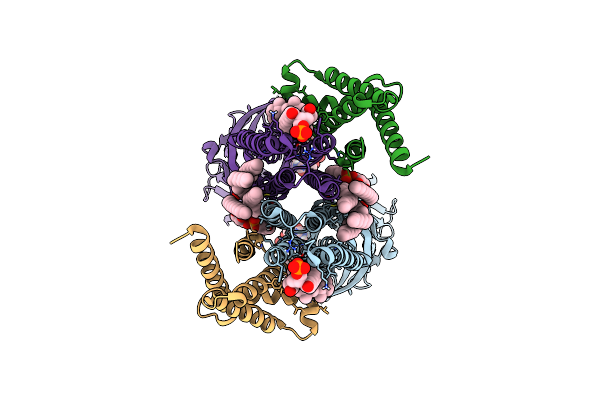 |
Cryo-Em Structure Of The Human Gabaa Receptor Alpha1 Subunit In Complex With The Assembly Factor Nacho/Tmem35A
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: MEMBRANE PROTEIN Ligands: PX6, PC7, PGT, CLR |
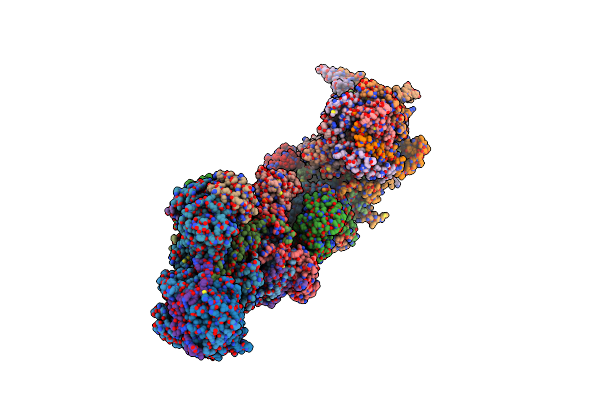 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: ELECTRON TRANSPORT Ligands: 3PE, SF4, FES, FMN, K, PC1, CDL, GTP, MG, NDP, ZN, EHZ, PGT, MYR |
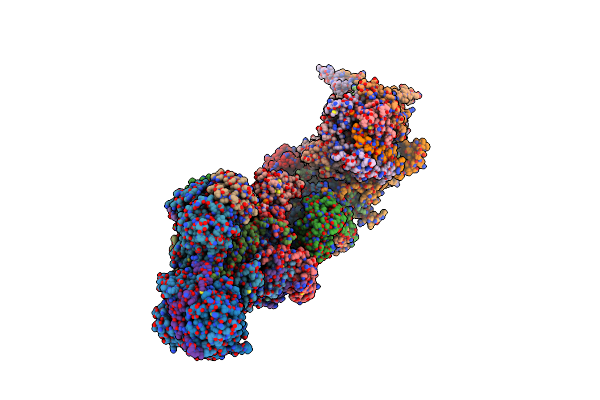 |
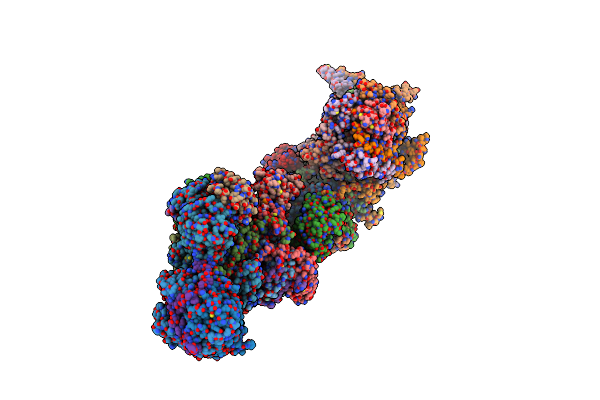
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: ELECTRON TRANSPORT Ligands: 3PE, SF4, FES, FMN, K, PC1, MYR, PGT, CDL, GTP, MG, NDP, ZN, EHZ |
 |
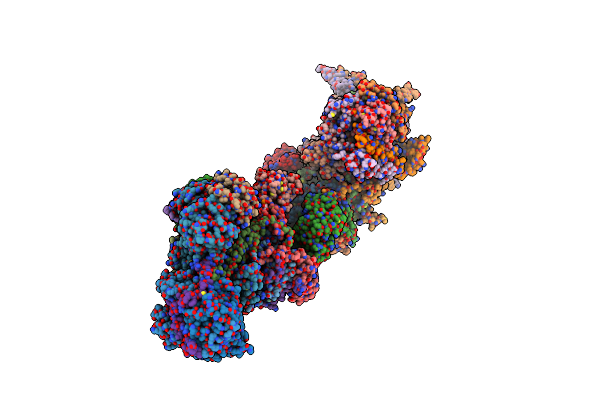
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: ELECTRON TRANSPORT Ligands: PC1, SF4, FES, FMN, K, 3PE, PGT, CDL, GTP, MG, NDP, ZN, EHZ, MYR |
 |
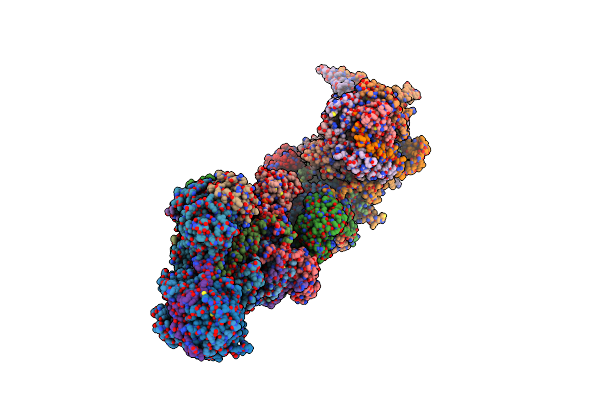
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: ELECTRON TRANSPORT Ligands: 3PE, SF4, FES, FMN, K, PC1, MYR, PGT, CDL, GTP, MG, NDP, ZN, EHZ |
 |
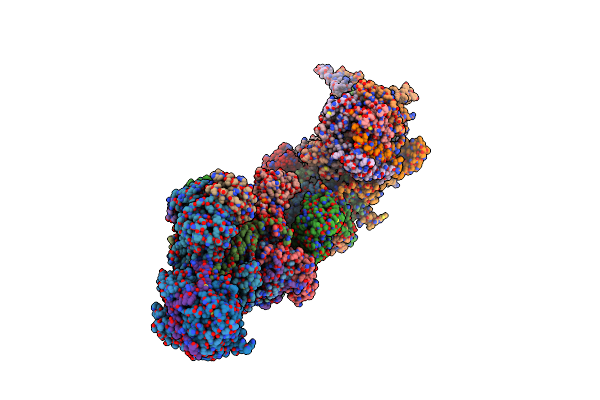
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: ELECTRON TRANSPORT Ligands: 3PE, SF4, FES, FMN, K, PC1, PGT, CDL, GTP, MG, NDP, ZN, EHZ, MYR |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: ELECTRON TRANSPORT Ligands: 3PE, SF4, PC1, FES, FMN, K, CDL, GTP, MG, NDP, ZN, EHZ, PGT, MYR |