Search Count: 2,243
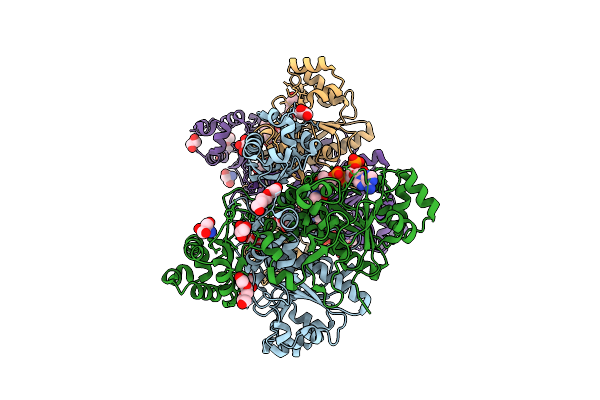 |
Succinyl-Coa:(R)-Benzylsuccinate Coa-Transferase (Bbsef), D178-Coa Adduct + Succinate (Weakly Occupied)
Organism: Aromatoleum aromaticum
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: GOL, PGE, COA, TRS, SIN |
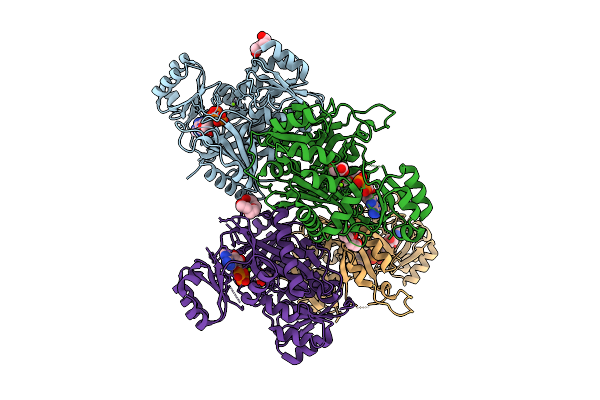 |
Crystal Structure Of Phosphoribosylaminoimidazole Carboxylase From Burkholderia Xenovorans (Atp Complex)
Organism: Paraburkholderia xenovorans lb400
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: ATP, PEG, MG, PGE, GOL, ADP, PG4 |
 |
Crystal Structure Of Phosphoribosylaminoimidazole Carboxylase From Burkholderia Xenovorans (Adp Complex)
Organism: Paraburkholderia xenovorans lb400
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: ADP, PEG, MG, PGE, GOL, PG4 |
 |
Crystal Structure Of Phosphoribosylaminoimidazole Carboxylase From Burkholderia Xenovorans (Amp Complex)
Organism: Paraburkholderia xenovorans lb400
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: AMP, PGE, MG, PEG, PG4, GOL |
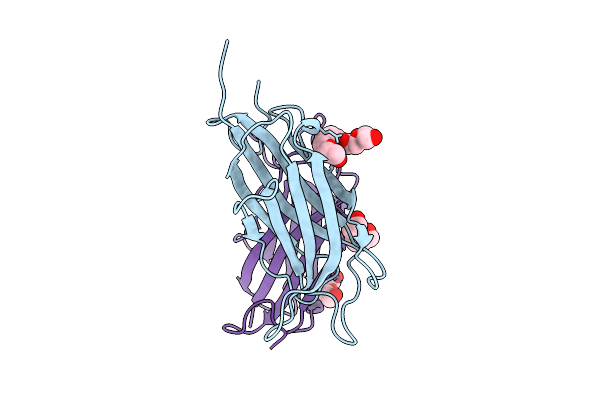 |
Crystal Structure Of A Calcium Bound C2 Domain Containing Protein From Trichomonas Vaginalis
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: 1PE, CL, PEG, PGE |
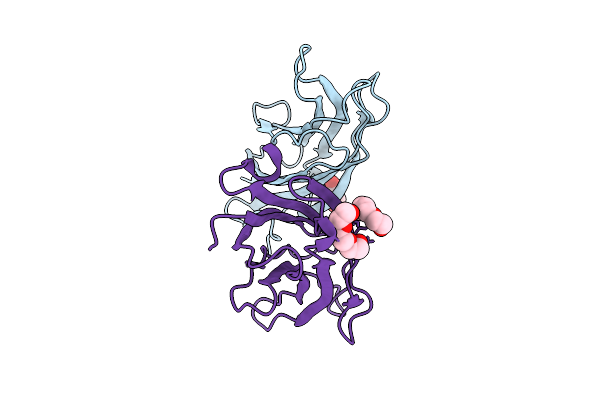 |
Crystal Structure Of A C2 Domain Containing Protein From Trichomonas Vaginalis
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: GOL, PGE |
 |
Structure Of Udp-Galactose-4-Epimerase (Gale) Bound To Fragment From Diamond Xchem Experiment.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: CARBOHYDRATE Ligands: NAD, JGA, MLI, PGE, EDO, CL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: DE NOVO PROTEIN Ligands: PGE, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: A1JLE, SO4, PG4, PGE, PEG |
 |
Mandh5 E303Q In Complex With Mannotetraose After Co-Crystalliztion With Mannotetraose At 1.6 Angstroms Resolution A Beta-D-Mannanase Of Gh5 Family From Dictyoglomus Thermophilium
Organism: Dictyoglomus thermophilum
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: HYDROLASE Ligands: BMA, MLT, PG4, PGE |
 |
Organism: Azotobacter vinelandii dj
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: METAL BINDING PROTEIN Ligands: SF4, S5Q, PEG, EDO, 1PE, PGE, MG, PG4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: IMMUNE SYSTEM Ligands: PGE, NA, PEG |
 |
Crystal Structure Of A Glyceraldehyde-3-Phosphate Dehydrogenase From Neisseria Gonorrhoeae In Complex With Nad And Glyceraldehyde-3-Phosphate
Organism: Neisseria gonorrhoeae nccp11945
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: G3H, PEG, NAD, NA, PGE, CL, GOL, EPE |
 |
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa And Inhibitor 20045
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, A1BHP, CL, PG4, 1PE, PGE |
 |
Crystal Structure Of The Nte Domain Of Suscdex (Bt3090) Dextran Transporter
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: NI, SCN, EDO, PEG, PGE |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSCRIPTION Ligands: ACT, PEG, CL, PGE, EDO |
 |
Crystal Structure Of A Glyceraldehyde-3-Phosphate Dehydrogenase From Neisseria Gonorrhoeae In Complex With Nad (P1 Form)
Organism: Neisseria gonorrhoeae nccp11945
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: OXIDOREDUCTASE Ligands: NAD, PGE |
 |
Organism: Thermomicrobium roseum dsm 5159
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: BG6, PEG, PG4, PGE, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN Ligands: HEM, GOL, A1ITR, DMS, CMO, ACY, SO4, PGE |
 |
Organism: Actinoplanes teichomyceticus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: BIOSYNTHETIC PROTEIN Ligands: LYS, SIN, PGE, VVO, CL, FLC, V |

