Search Count: 22
 |
Monomer Structure Of Transforming Growth Factor Beta Induced Protein (Tgfbip) G623R Fibril
Organism: Homo sapiens
Method: SOLID-STATE NMR Release Date: 2023-07-26 Classification: PROTEIN FIBRIL |
 |
Structure Of Transforming Growth Factor Beta Induced Protein (Tgfbip) G623R Fibril
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: PROTEIN FIBRIL |
 |
Organism: Megabalanus rosa
Method: SOLUTION NMR Release Date: 2020-01-15 Classification: STRUCTURAL PROTEIN |
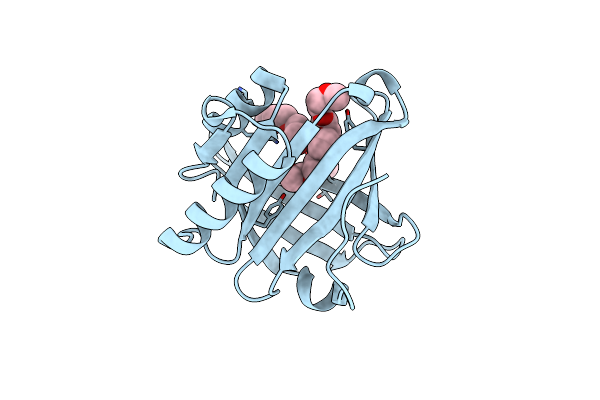 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-01-25 Classification: CHAPERONE Ligands: PG0, 1PG |
 |
 |
 |
Organism: Allamanda cathartica
Method: SOLUTION NMR Release Date: 2015-01-14 Classification: HYDROLASE INHIBITOR |
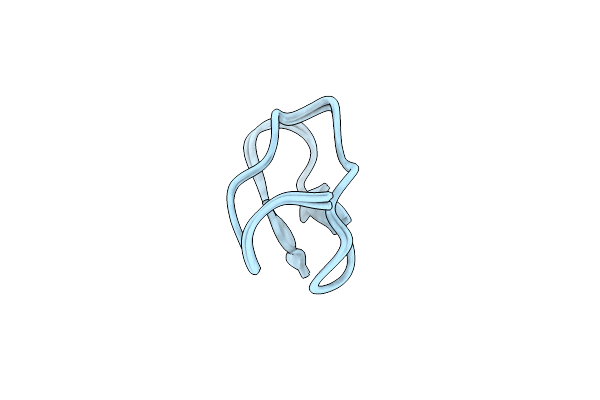 |
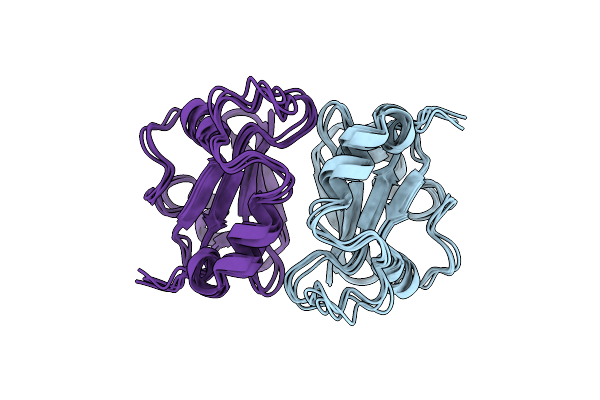
Organism: Allamanda cathartica
Method: SOLUTION NMR Release Date: 2015-01-14 Classification: HYDROLASE INHIBITOR |
 |
Organism: Geobacillus thermodenitrificans
Method: SOLUTION NMR Release Date: 2013-03-27 Classification: HYDROLASE |
 |
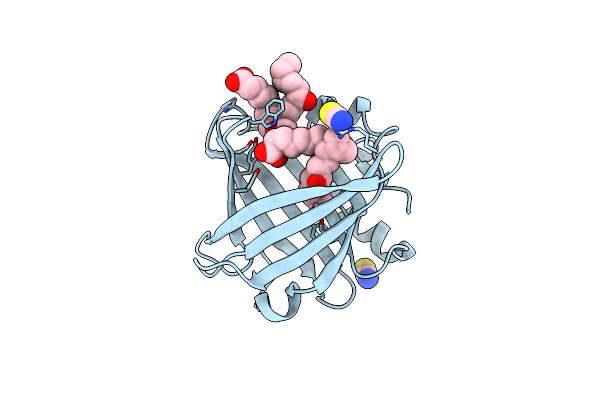
Crystal Structure Of Wild Type Human Lipocalin Pgds Bound With Peg Mme 2000
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2013-03-20 Classification: ISOMERASE Ligands: 1PG |
 |
Crystal Structure Of Wild Type Human Lipocalin Pgds In Complex With Substrate Analog U44069
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2013-03-20 Classification: ISOMERASE Ligands: PWZ, SCN |
 |
 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-01-12 Classification: ISOMERASE Ligands: CL, SCN |
 |
The Domain Features Of The Peripheral Stalk Subunit H Of The Methanogenic A1Ao Atp Synthase And The Nmr Solution Structure Of H1-47
Organism: Methanocaldococcus jannaschii
Method: SOLUTION NMR Release Date: 2009-07-21 Classification: STRUCTURAL PROTEIN |
 |
Nmr Solution Structure Of A Minimal Transmembrane Beta-Barrel Platform Protein
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2007-07-03 Classification: MEMBRANE PROTEIN |
 |
Organism: Methanocaldococcus jannaschii
Method: SOLUTION NMR Release Date: 2006-10-31 Classification: ISOMERASE Ligands: TSA |
 |
 |
Structural Role Of A Buried Salt Bridge In The 434 Repressor Dna-Binding Domain, Nmr, 20 Structures
Organism: Phage 434
Method: SOLUTION NMR Release Date: 1997-06-16 Classification: GENE REGULATING PROTEIN |
 |
Structural Role Of A Buried Salt Bridge In The 434 Repressor Dna-Binding Domain, Nmr, 20 Structures
Organism: Phage 434
Method: SOLUTION NMR Release Date: 1997-06-16 Classification: GENE REGULATING PROTEIN |

