Search Count: 4,063
All
Selected
 |
Crystal Structure Of The Double Mutant (Y128F And C222A) Of Sortase E From Thermobifida Fusca
Organism: Thermobifida fusca yx
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2026-03-04 Classification: HYDROLASE |
 |
Crystal Structure Of The Double Mutant (Y128A And C222A) Of Sortase E Mutant From Thermobifida Fusca
Organism: Thermobifida fusca yx
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2026-03-04 Classification: HYDROLASE |
 |
Organism: Thermobifida fusca yx
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2026-03-04 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-03-04 Classification: HYDROLASE Ligands: MG, ATP, ADP |
 |
Organism: Escherichia phage rb49
Method: SOLUTION NMR Release Date: 2026-02-25 Classification: ANTIMICROBIAL PROTEIN Ligands: ZN |
 |
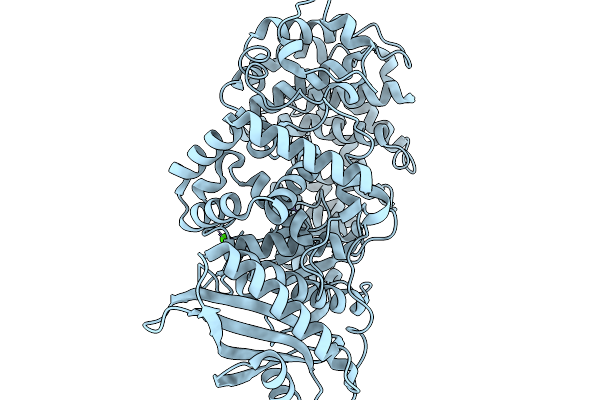
Organism: Paenibacillus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2026-02-18 Classification: HYDROLASE |
 |
Room Temperature Crystal Structure Of Collagenase Colh From Hathewaya Histolytica At 2.7 Angstrom Resolution
Organism: Hathewaya histolytica
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ZN, CA |
 |
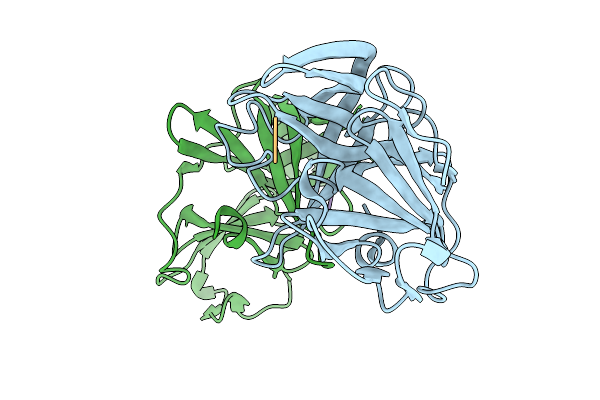
Organism: Porphyromonas gingivalis w83
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2026-01-14 Classification: METAL BINDING PROTEIN Ligands: ZN, CL |
 |
Organism: Norovirus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR |
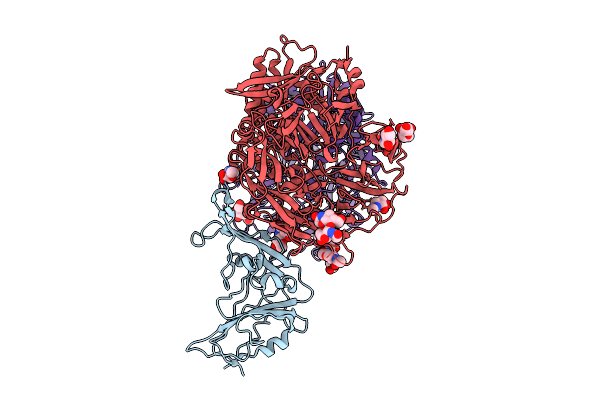 |
Organism: Middle east respiratory syndrome-related coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
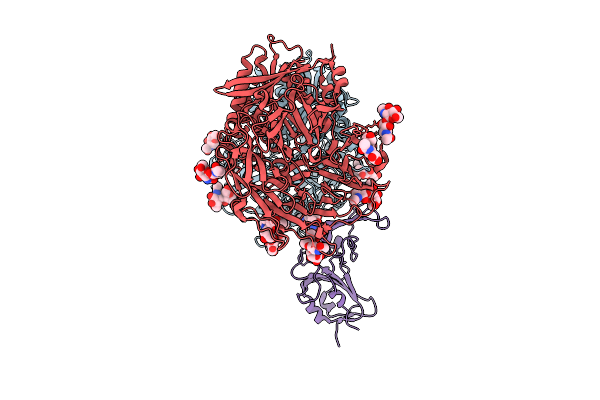 |
Organism: Homo sapiens, Bttp-betacov/gx2012
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
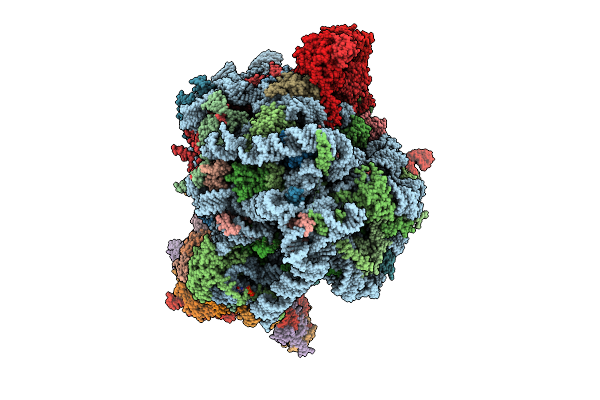 |
Human Quaternary Complex Of A Translating 80S Ribosome, Nac, Metap1 And Natd
Organism: Homo sapiens, Saccharomyces cerevisiae s288c, Aequorea victoria, Brachypodium distachyon
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: ZN, COA, GTP |
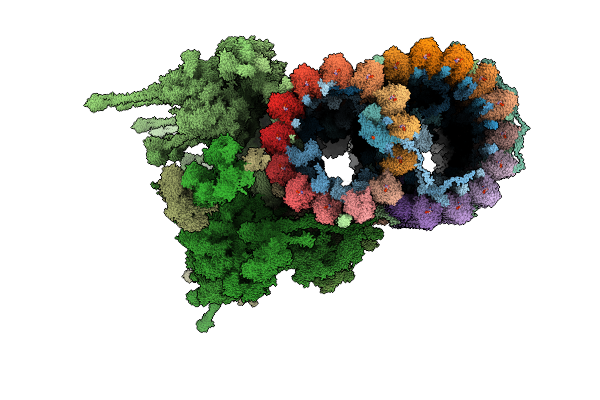 |
Organism: Leishmania tarentolae, Leishmania mexicana
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: STRUCTURAL PROTEIN Ligands: GDP, MG, GTP, ZN, CA, ATP |
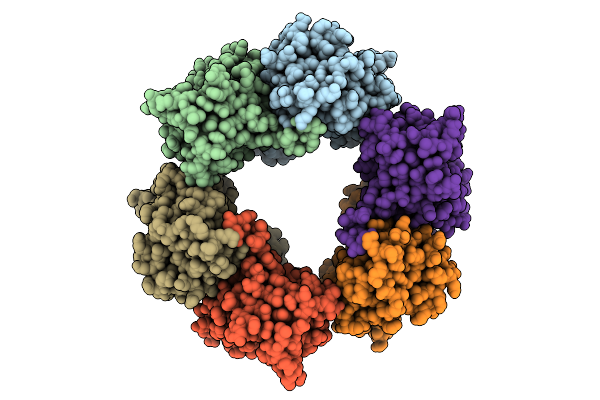 |
Cryo-Em Structure Of Candida Albicans Ph Regulated Antigen 1 (Pra1) Protein In The Absence Of Zn2+
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: METAL BINDING PROTEIN |
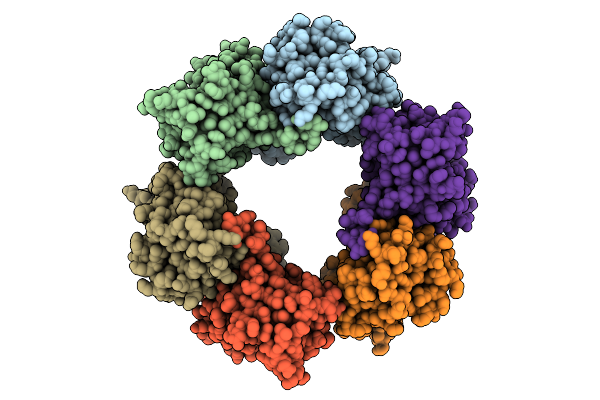 |
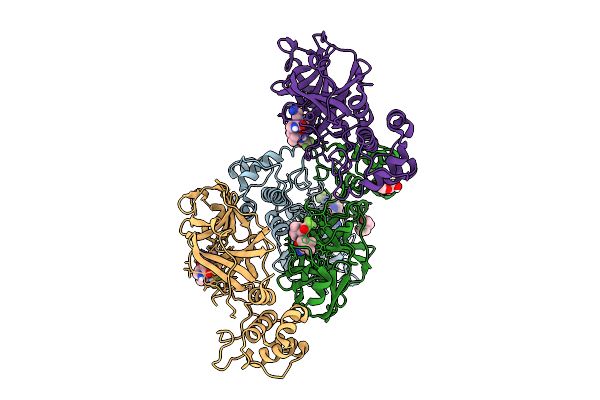
Cryo-Em Structure Of Candida Albicans Ph Regulated Antigen 1 (Pra1) Protein In Complex With Zn2+
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
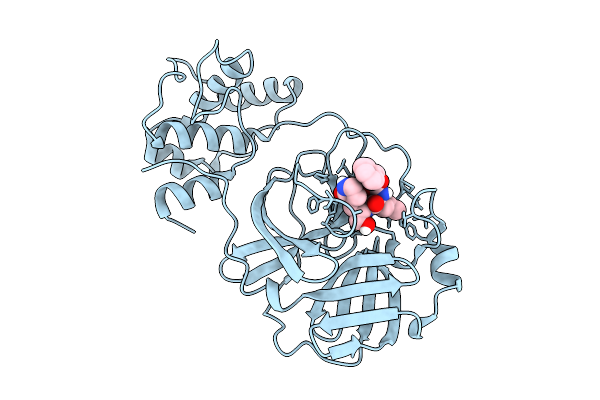
Co-Crystal Structure Of Feline Coronavirus Uu23 Main Protease With Nirmatrelvir
Organism: Feline coronavirus uu23
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: 4WI, PEG |
 |
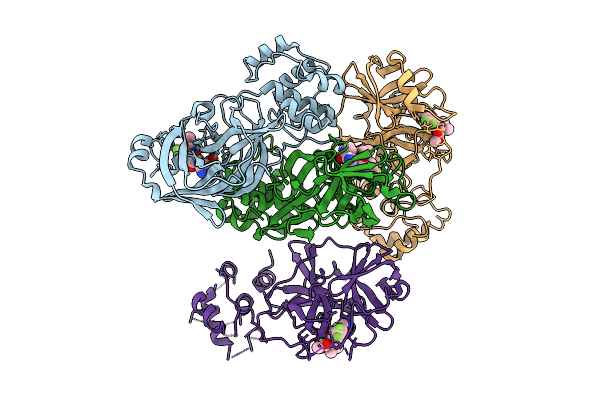
Organism: Feline coronavirus uu23
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: B1S |
 |
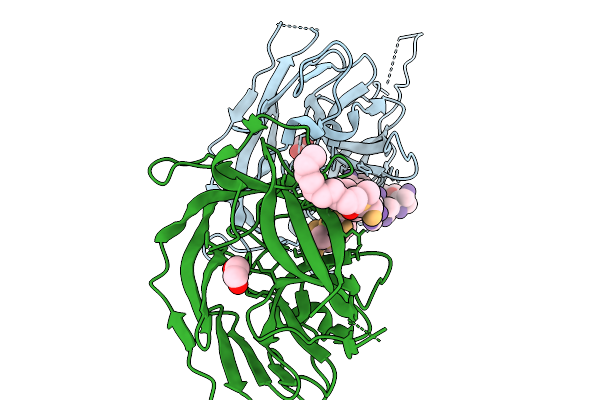
Co-Crystal Structure Of Feline Coronavirus Uu23 Main Protease With Ibuzatrelvir
Organism: Feline coronavirus uu23
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: YDL |
 |
Escherichia Coli Signal Peptidase I Delta 2-76 P84A In Complex With Lipopeptide Inhibitor
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2025-11-19 Classification: HYDROLASE/INHIBITOR Ligands: EDO, M12 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE Ligands: ADP |

