Search Count: 117
 |
Polaromonas Naphthalenivorans Phosphoenolpyruvate Carboxykinase In Complex With Pep (C2221)
Organism: Polaromonas naphthalenivorans
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2025-10-29 Classification: LYASE Ligands: MN, PEP |
 |
Polaromonas Naphthalenivorans Phosphoenolpyruvate Carboxykinase In Complex With Pep (P212121)
Organism: Polaromonas naphthalenivorans
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2025-10-29 Classification: LYASE Ligands: MN, PEP, PEG |
 |
Crystal Structure Of Pyrophosphate-Dependent Phosphofructokinase From Candidatus Prometheoarchaeum Syntrophicum With Phosphoenolpyruvate, Phosphate And Mg2+
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: PEP, PO4, MG |
 |
Crystal Structure Of Human Glucose-6-Phosphate Isomerase With Phosphoenol Pyruvate Ligand
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-03-12 Classification: ISOMERASE Ligands: PEP, PEG, PGE, TRS |
 |
Organism: Penaeus vannamei
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2023-12-13 Classification: LYASE Ligands: PEP, MG, SO4 |
 |
Crystal Structure Of Streptococcus Pneumoniae Pyruvate Kinase In Complex With Phosphoenolpyruvate
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2023-06-14 Classification: TRANSFERASE Ligands: MG, PEP, SO4 |
 |
Crystal Structure Of Streptococcus Pneumoniae Pyruvate Kinase In Complex With Phosphoenolpyruvate And Fructose 1,6-Bisphosphate
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-06-14 Classification: TRANSFERASE Ligands: FBP, PEP, MG, K |
 |
Dahp (3-Deoxy-D-Arabinoheptulosonate-7-Phosphate) Synthase Complexed With Mn(Ii), Pep, And Pi In Unbound:(Bound)2:Other Conformations
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-10-12 Classification: LYASE Ligands: MN, GAL, PEP, PO4 |
 |
Dahp (3-Deoxy-D-Arabinoheptulosonate-7-Phosphate) Synthase Complexed With Mn(Ii), Pep, And Pi In Unbound:(Bound)2:Other Conformations
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2022-10-12 Classification: LYASE Ligands: MN, PEP, PO4 |
 |
Aspergillus Fumigatus Enolase Bound To Phosphoenolpyruvate And 2-Phosphoglycerate
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-03-16 Classification: LYASE Ligands: MG, 2PG, PEP |
 |
Mycobacterium Tuberculosis Enolase Mutant - E204A Complex With Phosphoenolpyruvate
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: PEP, PEG, EDO, ACT, MG |
 |
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2022-02-16 Classification: METAL BINDING PROTEIN Ligands: PEP, MG |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-12-01 Classification: LYASE Ligands: PEP, PEG, EDO, ACT, MG, CL |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2021-10-13 Classification: LYASE Ligands: MN, NA, PEP, GDP, CO2 |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2021-10-13 Classification: LYASE Ligands: CO2, GDP, PEP, MN |
 |
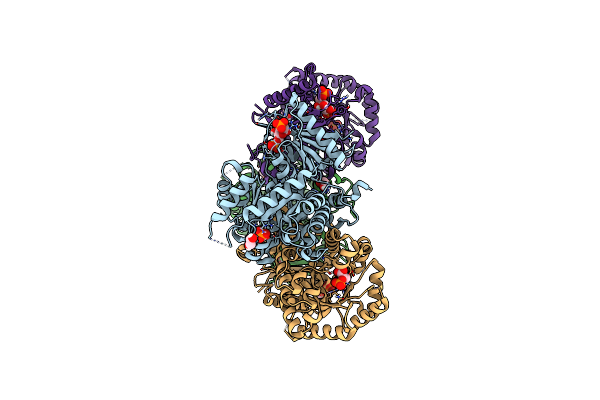
Ambient Temperature Structure Of Bifidobacgterium Longum Phosphoketolase With Thiamine Diphosphate And Phosphoenol Pyuruvate
Organism: Bifidobacterium longum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-06-02 Classification: LYASE Ligands: PEP, TPP, CA |
 |
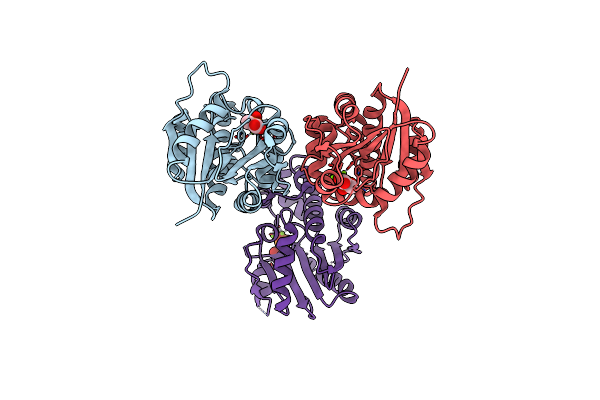
The Structure Of Human Liver Pyruvate Kinase, Hlpyk-D499N, In Complex With Fru-1,6-Bp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-06-19 Classification: TRANSFERASE Ligands: FBP, EDO, PEP |
 |
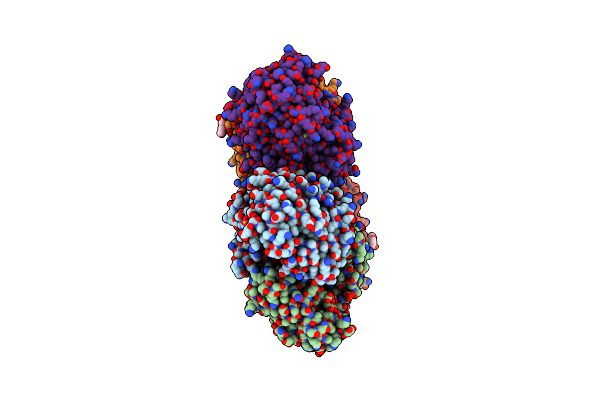
Crystal Structure Of Mycoibacterium Tuberculosis Rv2983 In Complex With Pep
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2018-12-19 Classification: TRANSFERASE Ligands: PEP, MG |
 |
Crystal Structure Of Enolase From E. Coli With A Mixture Of Apo Form, Substrate, And Product Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2018-10-31 Classification: LYASE Ligands: MG, SO4, GOL, PEP, 2PG, MES |
 |
The Structure Of A Dimeric Type Ii Dah7Ps Associated With Pyocyanin Biosynthesis In Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-10-03 Classification: TRANSFERASE Ligands: PEP, CO, CL |

