Search Count: 54
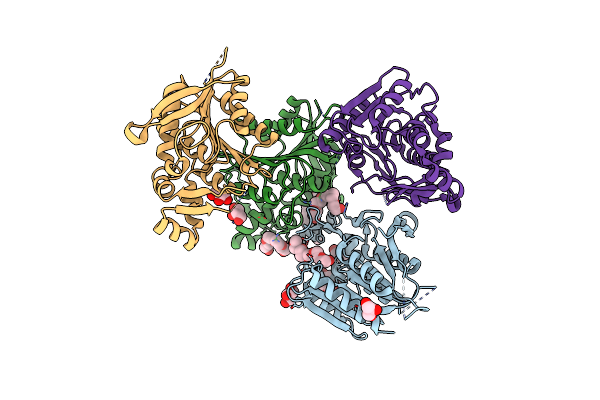 |
Crystal Structure Of Sm0281, An Alpha/Beta Hydrolase From Sinorhizobium Meliloti
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2025-02-12 Classification: HYDROLASE Ligands: GOL, PE3, CL |
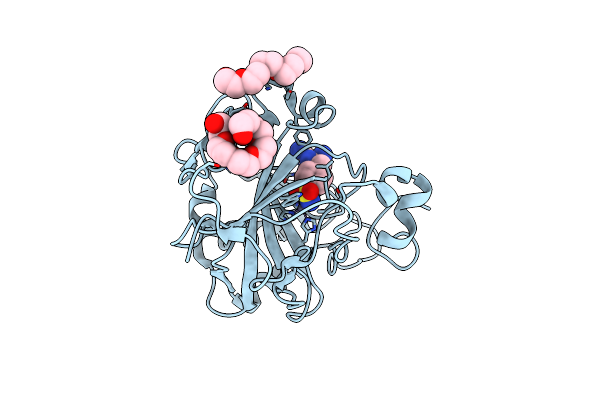 |
Human Carbonic Anhydrase Ii In Complex With Biguanide Derivative Inhibitor 1-Carbamimidamido-N-[(4 Sulfamoylphenyl)Methyl]Methanimidamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2025-01-29 Classification: LYASE Ligands: ZN, A1H15, PE3, 1PE |
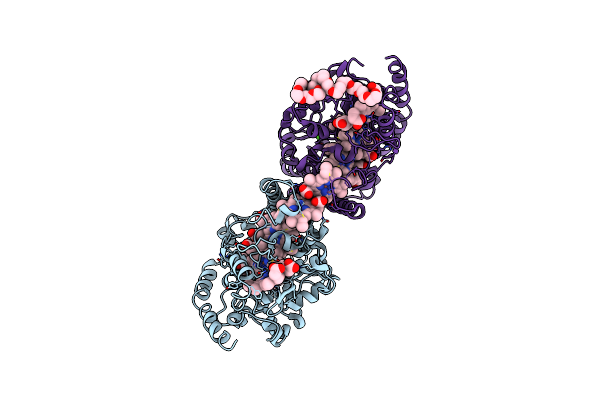 |
Organism: Shewanella oneidensis
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2025-01-15 Classification: ELECTRON TRANSPORT Ligands: HEC, CA, PE3 |
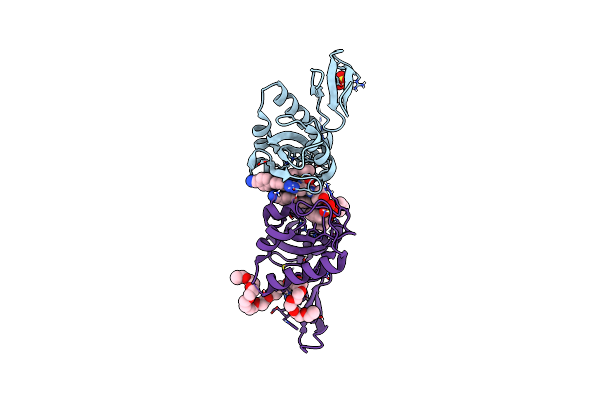 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2024-09-25 Classification: ISOMERASE Ligands: A1D9K, SO4, PE3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2024-05-29 Classification: CELL CYCLE Ligands: PE3, PEG, YPK |
 |
Crystal Structure Of A Gh12-2 Family Cellulase From Thermococcus Sp. 2319X1
Organism: Thermococcus sp. 2319x1
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2022-08-24 Classification: HYDROLASE Ligands: GOL, PE3, CA, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2020-08-19 Classification: TRANSCRIPTION/INHIBITOR Ligands: IOD, GOL, NH2, PE3 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-02-05 Classification: IMMUNE SYSTEM Ligands: 1PE, TOE, PE3, PG4 |
 |
Crystal Structure Of The Gh26 Domain From Pbgh26-Gh5A Endo-Beta-Mannanase/Endo-Beta-Glucanase From Prevotella Bryantii
Organism: Prevotella bryantii b14
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2019-10-16 Classification: HYDROLASE Ligands: GOL, PE3, SO4 |
 |
Organism: Simiduia agarivorans (strain dsm 21679 / jcm 13881 / bcrc 17597 / sa1)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-07-31 Classification: HYDROLASE Ligands: ZN, ACT, GOL, PE3, CL |
 |
Crystal Structure Of The Caldicellulosiruptor Lactoaceticus Gh74 (Clgh74A) Enzyme In Complex With Llg Xyloglucan
Organism: Caldicellulosiruptor lactoaceticus 6a
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2019-07-31 Classification: HYDROLASE Ligands: PE3, GOL, TRS |
 |
Crystal Structure Of Peroxisome Proliferator-Activated Receptor Delta (Ppard)Lbd In Complex With Dn003316
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-07-10 Classification: STRUCTURAL PROTEIN Ligands: 9RF, B7G, PE3 |
 |
Crystal Structure Of The Catalytic Domain From Gh74 Enzyme Pogh74 From Paenibacillus Odorifer, Apoenzyme
Organism: Paenibacillus odorifer
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-23 Classification: HYDROLASE Ligands: MG, PE3, GOL, PO4, CL |
 |
Crystal Structure Of The Catalytic Domain From Gh74 Enzyme Pogh74 From Paenibacillus Odorifer, In Complex With Xlx Xyloglucan
Organism: Paenibacillus odorifer
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-01-23 Classification: HYDROLASE Ligands: GOL, PE3, CL |
 |
Crystal Structure Of The Catalytic Domain From Gh74 Enzyme Pogh74 From Paenibacillus Odorifer, D60A Mutant In Complex With Xxlg And Xgxxlg Xyloglucan
Organism: Paenibacillus odorifer
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-01-23 Classification: HYDROLASE Ligands: CL, MG, PE3, GOL |
 |
Crystal Structure Of Meta-Aac0038, An Environmental Aminoglycoside Resistance Enzyme, H168A Mutant In Complex With Acetyl-Coa
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-10-24 Classification: TRANSFERASE Ligands: ACO, CL, GOL, SO4, PE3 |
 |
Crystal Structure Of Meta-Aac0038, An Environmental Aminoglycoside Resistance Enzyme, Mutant H168A In Abortive Complex With Gentamicin-Coa
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-10-24 Classification: TRANSFERASE/ANTIBIOTIC Ligands: COA, LLL, CL, SO4, PE3, GOL |
 |
Crystal Structure Of Aminoglycoside Acetyltransferase Aac(3)-Iva, H154A Mutant, In Complex With Gentamicin C1A
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2018-10-24 Classification: TRANSFERASE/ANTIBIOTIC Ligands: LLL, ZN, EPE, GOL, EDO, PE3 |
 |
Crystal Structure Of The Bc1960 Peptidoglycan N-Acetylglucosamine Deacetylase In Complex With 4-Naphthalen-1-Yl-~{N}-Oxidanyl-Benzamide
Organism: Bacillus cereus (strain atcc 14579 / dsm 31 / jcm 2152 / nbrc 15305 / ncimb 9373 / nrrl b-3711)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-06-20 Classification: HYDROLASE Ligands: SO4, 5YA, PE3, PEG, PG0, NA, CL, ZN, DMS |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-03-28 Classification: TRANSFERASE Ligands: PE3, SO4 |

