Search Count: 68
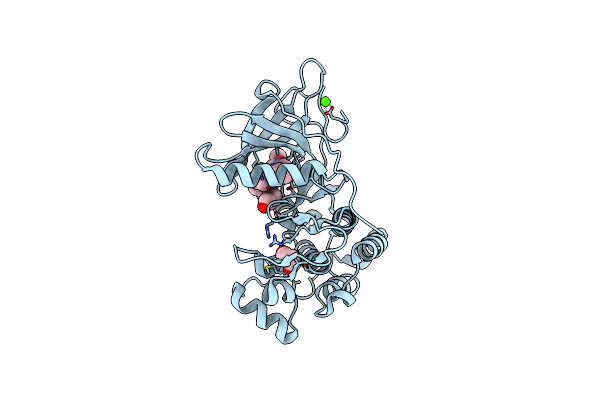 |
Co-Crystal Structure Of The Kinase Domain Of Egfr With Non-Covalent Osimertinib
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TRANSFERASE Ligands: PDO, Q6K, CA |
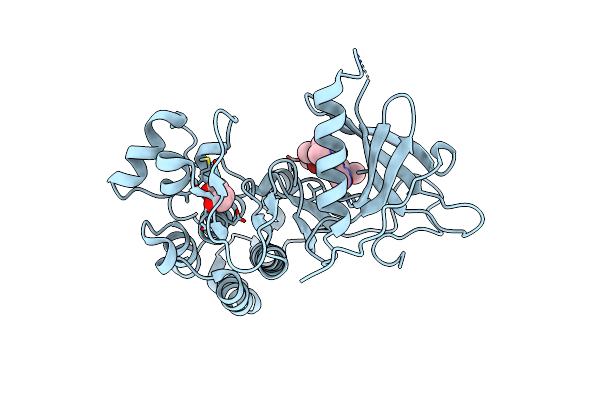 |
Crystal Structure Of The Kinase Domain Of Egfr Soaked With Non-Covalent Osimertinib
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TRANSFERASE Ligands: PDO, Q6K |
 |
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: HYDROLASE Ligands: A1EBP, GOL, BEZ, PDO, BU1, PGO |
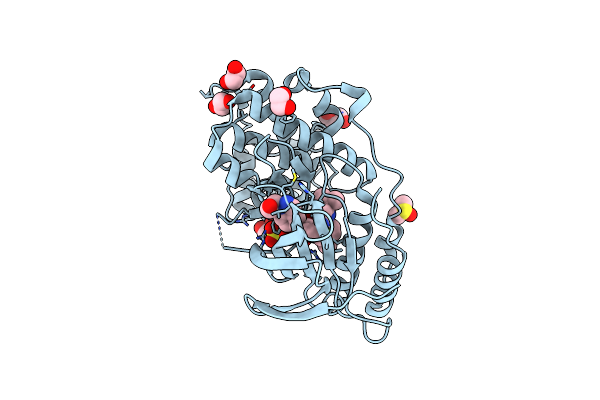 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-11-13 Classification: TRANSFERASE Ligands: SO4, EDO, GOL, DMS, PDO, A1AWV |
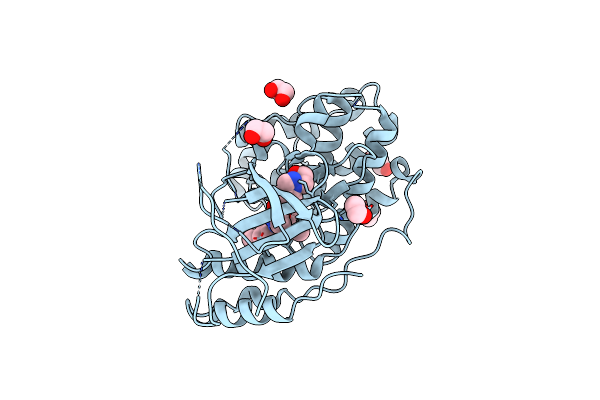 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2024-11-13 Classification: TRANSFERASE Ligands: EDO, A1AWV, PDO |
 |
The Crystal Structure Of Protein A21, A Component Of The Conserved Poxvirus Entry-Fusion Complex
Organism: Vaccinia virus western reserve
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-09-04 Classification: VIRAL PROTEIN Ligands: EOH, EDO, MOH, IOD, CL, PDO, PGR, NA, PG0, DHL, PGE, PEG, POL, GOL, IPA |
 |
Structure Of A Salivary Alpha-Glucosidase From The Mosquito Vector Aedes Aegypti.
Organism: Aedes aegypti
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2024-03-20 Classification: HYDROLASE Ligands: NAG, CYS, GOL, TRS, IPA, PDO, CA, CL |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-09-06 Classification: TRANSFERASE Ligands: EDO, SO4, MES, PO4, PDO, 627 |
 |
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: PDO |
 |
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: HEM, PGO, BU1, PDO |
 |
Crystal Structure Of Pseudomonas Aeruginosa S-Adenosyl-L-Homocysteine Hydrolase Inhibited By Cd2+ Ions
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: NAD, ADN, BU1, PDO, K, CD, HEZ, CL |
 |
Crystal Structure Of Pseudomonas Aeruginosa S-Adenosyl-L-Homocysteine Hydrolase Inhibited By Hg2+ Ions
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: NAD, ADN, BU1, PDO, K, HG, CL |
 |
Crystal Structure Of Pseudomonas Aeruginosa S-Adenosyl-L-Homocysteine Hydrolase Inhibited By Co2+ Ions.
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: NAD, ADN, PDO, K, CO, BU1, HEZ, CL |
 |
Crystal Structure Of Pseudomonas Aeruginosa S-Adenosyl-L-Homocysteine Hydrolase Inhibited By Zn2+ Ions
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: NAD, ADN, BU1, PG4, K, ZN, BCN, HEZ, PDO, CL |
 |
Epstein-Barr Virus Encoded Apoptosis Regulator Bhrf1 In Complex With Puma Bh3
Organism: Epstein-barr virus (strain b95-8), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-08-10 Classification: APOPTOSIS Ligands: PO4, EDO, NH4, PDO, MG, CL |
 |
Catalytic Domain Of Udp-Glucose Glycoprotein Glucosyltransferase From Chaetomium Thermophilum In Complex With Udp
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2022-06-22 Classification: TRANSFERASE Ligands: UDP, NAG, CA, PDO, EDO |
 |
Catalytic Domain Of Udp-Glucose Glycoprotein Glucosyltransferase From Chaetomium Thermophilum In Complex With Udp-2-Deoxy-2-Fluoro-D-Glucose
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-06-22 Classification: TRANSFERASE Ligands: NAG, U2F, CA, PDO |
 |
Catalytic Domain Of Udp-Glucose Glycoprotein Glucosyltransferase From Chaetomium Thermophilum In Complex With Udp-Glucose (Conformation 2)
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-04-20 Classification: TRANSFERASE Ligands: UPG, CA, IPA, PDO |
 |
Crystal Structure Of Erap2 Aminopeptidase In Complex With Phosphinic Pseudotripeptide((1R)-1-Amino-3-Phenylpropyl){(2S)-3-[((2S)-1-Amino-1-Oxo-3-Phenylpropan-2-Yl)Amino]-2-{[3-(2-Hydroxyphenyl)-Isoxazol-5-Yl]Methyl}-3-Oxopropyl}Phosphinic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-01-26 Classification: HYDROLASE Ligands: 62S, NAG, PEG, PGE, MES, EDO, PDO, IMD, ZN |
 |
Crystal Structure Of Metal-Citrate-Binding Mutant (D28A) Protein (Mcta) Of Abc Transporter Endogenously Bound To Citrate
Organism: Thermus thermophilus (strain atcc 27634 / dsm 579 / hb8)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-12-01 Classification: TRANSPORT PROTEIN Ligands: CIT, EDO, PDO, PGR, CO2 |

