Search Count: 15
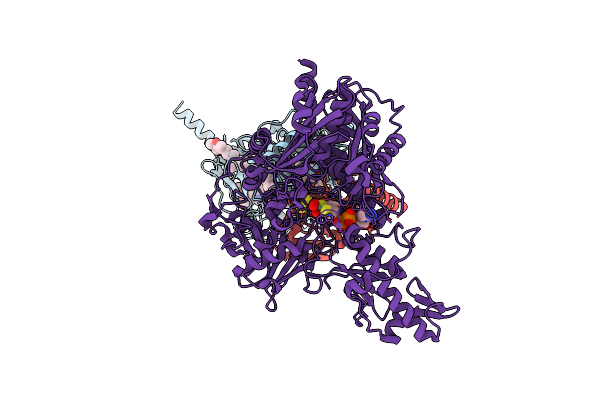 |
Cryo-Em Structure Of Membrane-Bound Aldehyde Dehydrogenase From Gluconobacter Oxydans
Organism: Gluconobacter oxydans
Method: ELECTRON MICROSCOPY Release Date: 2023-08-02 Classification: OXIDOREDUCTASE Ligands: HEC, U10, FES, PCD |
 |
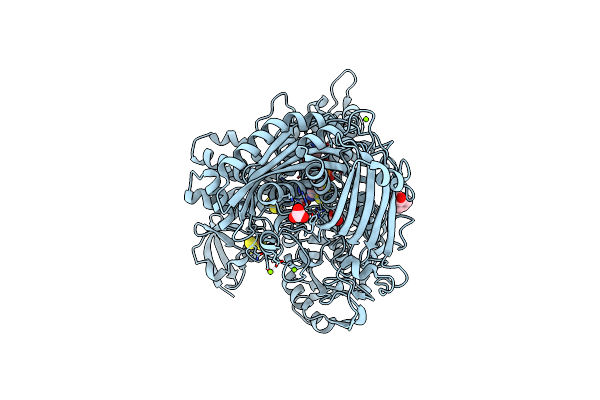
Aldehyde Oxidoreductase From Desulfovibrio Gigas (Mop), Soaked With Benzaldehyde
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2014-10-08 Classification: OXIDOREDUCTASE Ligands: FES, PGE, IPA, BCT, MG, CL, PCD, HBX |
 |
Aldehyde Oxidoreductase From Desulfovibrio Gigas (Mop), Soaked With 3- Phenylpropionaldehyde
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-10-08 Classification: OXIDOREDUCTASE Ligands: FES, 3PL, BCT, MG, CL, PCD |
 |
Aldehyde Oxidoreductase From Desulfovibrio Gigas (Mop), Soaked With Trans-Cinnamaldehyde
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2014-10-08 Classification: OXIDOREDUCTASE Ligands: FES, IPA, BCT, MG, CL, PCD, HCI |
 |
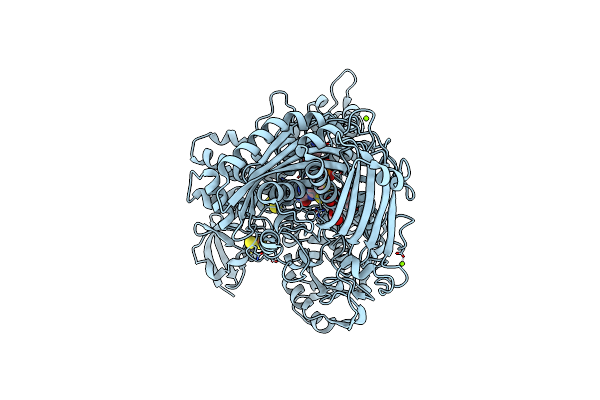
Aldehyde Oxidoreductase From Desulfovibrio Gigas (Mop), Soaked With Sodium Dithionite And Sodium Sulfide
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2014-01-15 Classification: OXIDOREDUCTASE Ligands: FES, IPA, BCT, MG, CL, PCD, PEO |
 |
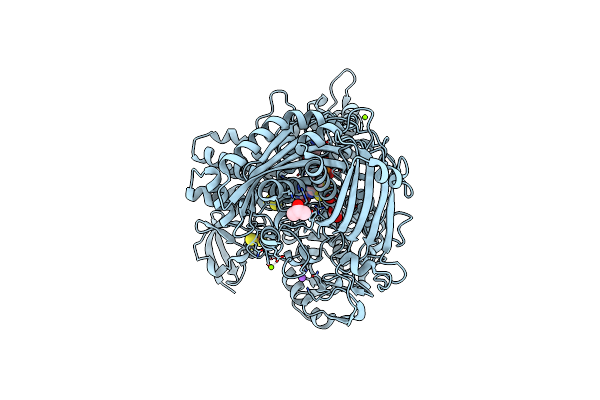
Aldehyde Oxidoreductase From Desulfovibrio Gigas (Mop), Activated With Sodium Dithionite And Sodium Sulfide
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2014-01-15 Classification: OXIDOREDUCTASE Ligands: MG, FES, IPA, BCT, CL, PCD, PEO |
 |
Aldehyde Oxidoreductase From Desulfovibrio Gigas (Mop), Soaked With Hydrogen Peroxide
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-01-15 Classification: OXIDOREDUCTASE Ligands: FES, IPA, BCT, NA, MG, CL, PCD, PEO |
 |
Crystal Structure Of The Aldehyde Dehydrogenase (A.K.A. Aor Or Mop) Of Desulfovibrio Gigas Covalently Bound To [Aso3]-
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2010-02-16 Classification: OXIDOREDUCTASE Ligands: FES, MG, LI, CL, URE, PCD, AST, IPA, CA |
 |
Glycerol Inhibited Form Of Aldehyde Oxidoreductase From Desulfovibrio Gigas
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2009-10-06 Classification: OXIDOREDUCTASE Ligands: MG, CL, FES, PCD, GOL |
 |
Ethylene Glycol Inhibited Form Of Aldehyde Oxidoreductase From Desulfovibrio Gigas
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2009-10-06 Classification: OXIDOREDUCTASE Ligands: MG, CL, FES, PCD, EDO |
 |
Organism: Thauera aromatica
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2004-12-21 Classification: OXIDOREDUCTASE Ligands: CL, NA, PCD, EPE, FAD, SF4, FES, K, SO4 |
 |
Organism: Thauera aromatica
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-12-21 Classification: OXIDOREDUCTASE Ligands: PCD, EPE, FAD, SF4, FES, SO4 |
 |
Crystal Structure Of The Aldehyde Dehydrogenase (A.K.A. Aor Or Mop) Of Desulfovibrio Gigas Covalently Bound To [Aso3]-
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-07-27 Classification: OXIDOREDUCTASE Ligands: CL, MG, PCD, AST, FES |
 |
Structure Refinement Of The Aldehyde Oxidoreductase From Desulfovibrio Gigas At 1.28 A
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2004-07-27 Classification: OXIDOREDUCTASE Ligands: CL, MG, PCD, FES, IPA |
 |
Organism: Hydrogenophaga pseudoflava
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2000-09-15 Classification: HYDROLASE Ligands: FES, PCD, FAD |

