Search Count: 6
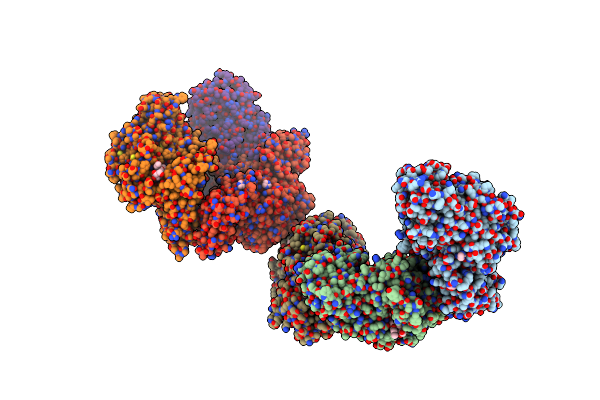 |
Crystal Structure Of Susa Amylase From Bacteroides Thetaiotaomicron Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, OC9, IMD, CA |
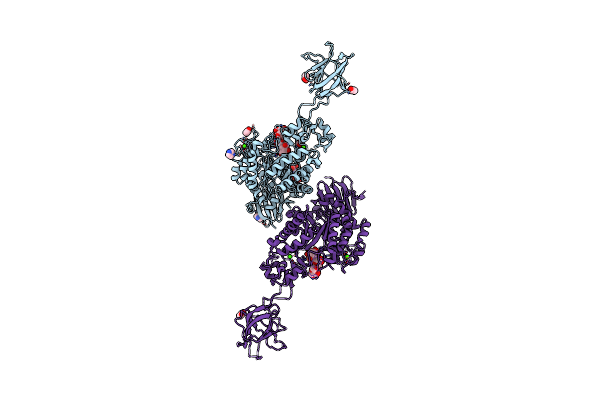 |
Crystal Structure Of Susg From Bacteroides Thetaiotaomicron Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, OC9, A1ILG, CA, IMD, ACT |
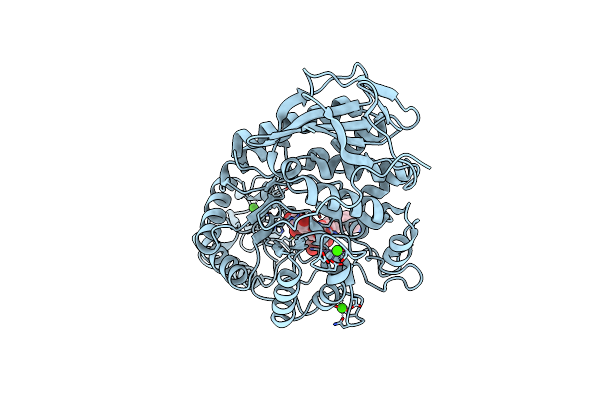 |
Crystal Structure Of Amylase 5 (Amy5) From Ruminococcus Bromii Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Ruminococcus bromii
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, A1IHI, CA |
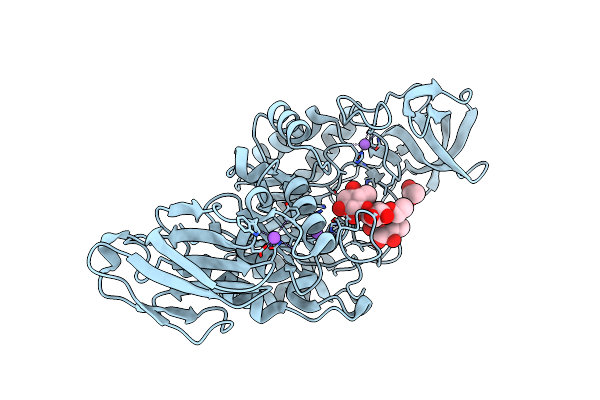 |
Crystal Structure Of K38 Amylase From Bacillus Sp. Strain Ksm-K38 Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Bacillus sp. ksm-k38
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, OC9, ACT, NA |
 |
Structure Of The D125N Mutant Of The Catalytic Domain Of The Bacillus Circulans Alpha-1,6 Mannanase In Complex With An Alpha-1,6-Alpha-Manno-Cyclophellitol Trisaccharide Inhibitor
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-04-28 Classification: HYDROLASE Ligands: PBW, EDO |
 |
Structure Of The Catalytic Domain Of The Bacillus Circulans Alpha-1,6 Mannanase In Complex With An Alpha-1,6- Alpha-Manno-Cyclophellitol Carbasugar-Stabilised Trisaccharide Inhibitor
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2021-04-28 Classification: HYDROLASE Ligands: EDO, PBW, M96, MAN |

