Search Count: 16
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.60 Å Release Date: 2025-01-08 Classification: MEMBRANE PROTEIN Ligands: CA, PAB |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.67 Å Release Date: 2025-01-08 Classification: MEMBRANE PROTEIN Ligands: CA, PAB, CL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-08 Classification: MEMBRANE PROTEIN Ligands: CA, PAB, CL |
 |
Crystal Structure Of Sulfonamide Resistance Enzyme Sul2 In Complex With 7,8-Dihydropteroate, Magnesium, And Pyrophosphate
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: 78H, POP, MG, GOL, CL, PAB |
 |
Crystal Structure Of Sulfonamide Resistance Enzyme Sul3 In Complex With Reaction Intermediate
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: XHP, PAB, GOL, CL, MG, POP |
 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 5.5 In Complex With 4-Aminobenzoic Acid
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-02-15 Classification: TRANSFERASE Ligands: PAB, BTB, CL |
 |
Structure Of Mouse Bai1 (Adgrb1) In Complex With Mouse Nogo Receptor (Rtn4R)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-11-10 Classification: CELL ADHESION Ligands: NAG, PAB, PG4, EDO, CL, ACT, MAN |
 |
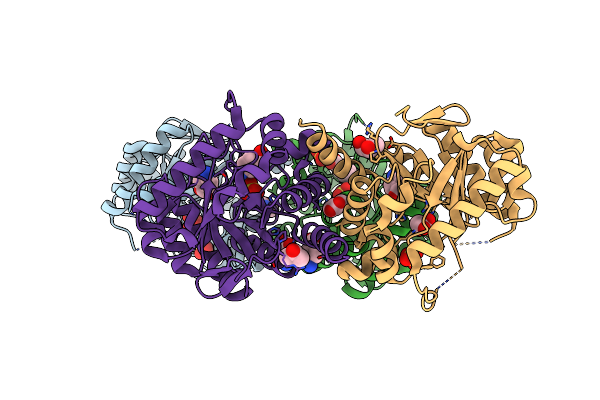
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-08-29 Classification: TRANSFERASE Ligands: HH2, APC, PAB, MG, PE0, 5AD |
 |
Xanthomonas Albilineans Dihydropteroate Synthase With 4-Aminobenzoic Acid At 1.65 A
Organism: Xanthomonas albilineans gpe pc73
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-06-06 Classification: TRANSFERASE Ligands: PAB, SO4, GOL |
 |
Crystal Structure Of The Yersinia Pestis Dihydropteroate Synthetase With Substrate Transition State Complex.
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2012-03-14 Classification: TRANSFERASE/TRANSFERASE SUBSTRATE Ligands: XHP, POP, PAB, MG |
 |
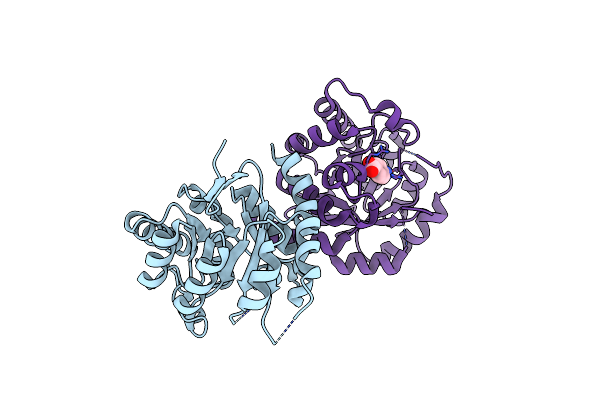
Organism: Pseudomonas mendocina
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2011-12-21 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: FE, PAB, P6G, ETE |
 |
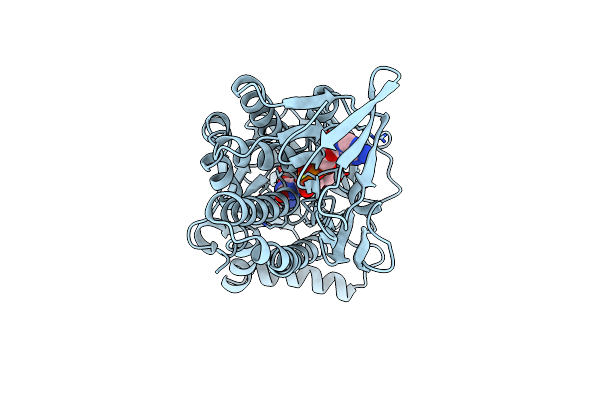
Crystal Structure Of Dihydropteroate Synthase From Thermus Thermophilus Hb8 In Complex With 4-Aminobenzoate
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-03-27 Classification: TRANSFERASE Ligands: PAB |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1996-04-03 Classification: OXIDOREDUCTASE Ligands: FAD, PAB |
 |
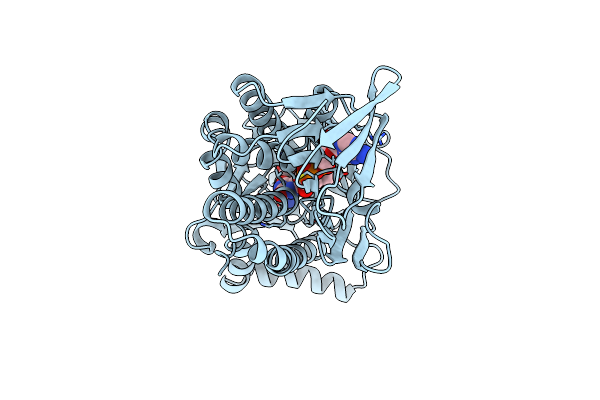
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1996-04-03 Classification: OXIDOREDUCTASE Ligands: FAD, PAB |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1996-04-03 Classification: OXIDOREDUCTASE Ligands: FAD, PAB |
 |
Crystal Structures Of Wild-Type P-Hydroxybenzoate Hydroxylase Complexed With 4-Aminobenzoate, 2,4-Dihydroxybenzoate And 2-Hydroxy-4-Aminobenzoate And Of The Try222Ala Mutant, Complexed With 2-Hydroxy-4-Aminobenzoate. Evidence For A Proton Channel And A New Binding Mode Of The Flavin Ring
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1994-09-30 Classification: OXIDOREDUCTASE Ligands: FAD, PAB |

