Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Photoactivation In Bacteriophytochrome, High Resolution Cryo Structure In The Dark.
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, P33 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: ISOMERASE Ligands: A1EDZ, P33, SO4, CL |
 |
Organism: Anopheles gambiae
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: GOL, PEG, EDO, P33, PO4, XPE, P6G, PGE, PG4 |
 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: MO, MGD, F3S, EDO, PEG, SBO, P33, PGE, O, NA, FES, GOL, 4MO, P4G, 1PE, PG0 |
 |
Organism: Pseudorhizobium banfieldiae
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: SER, 4MO, F3S, EDO, PEG, MGD, SO4, P33, GOL, O, FES, 1PE |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: ANTIBIOTIC Ligands: DMS, SO4, PE5, EDO, PEG, GOL, CL, NA, P33 |
 |
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-04-02 Classification: ISOMERASE Ligands: SO4, GOL, P33, PG4 |
 |
Human Angiotensin-1 Converting Enzyme N-Domain In Complex With A Diprolyl Inhibitor- Sg15
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: NAG, P33, PEG, ZN, CL, A1IJZ, PG4, MG |
 |
Human Angiotensin-1 Converting Enzyme N-Domain In Complex With A Diprolyl Inhibitor- Sg17
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: NAG, ZN, CL, A1IJX, PGE, P33, PG4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2025-01-22 Classification: DE NOVO PROTEIN Ligands: FMT, NA, GOL, NH4, P33 |
 |
Structure Of The Fast1-Fast2-Rap Module From Human Fastkd4 By Carrier-Driven Crystallisation With Maltose Binding Protein From E. Coli.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-01-15 Classification: RNA BINDING PROTEIN Ligands: P33, PO4, GLC, FRU |
 |
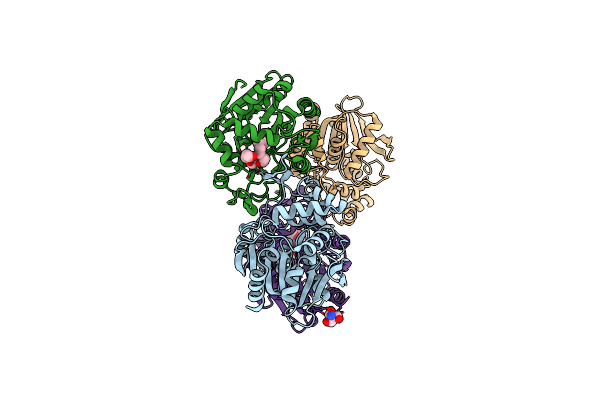
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: P33, BTB |
 |
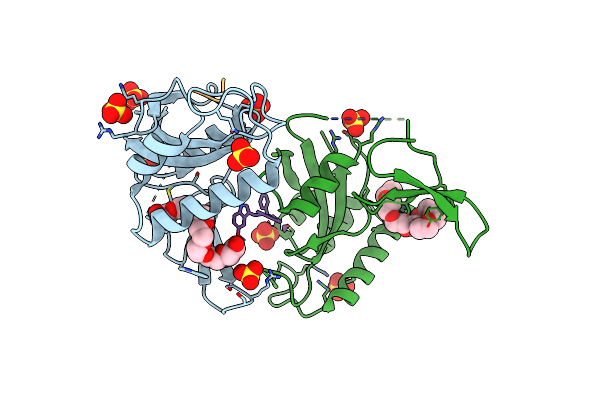
S102A Mutant Of Poly(3-Hydroxybutyrate) Depolymerase Phaz From Bacillus Thuringiensis
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: P33, TRS |
 |
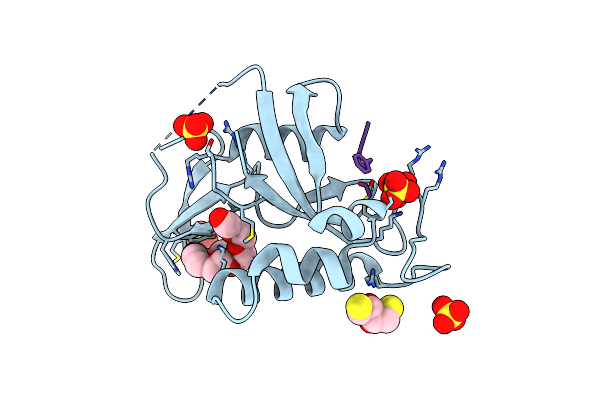
Human R14A Pin1 Covalently Bound To Inhibitor 158F10 In P21 21 21 Space Group
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2024-11-06 Classification: ISOMERASE/INHIBITOR Ligands: P33, SO4 |
 |
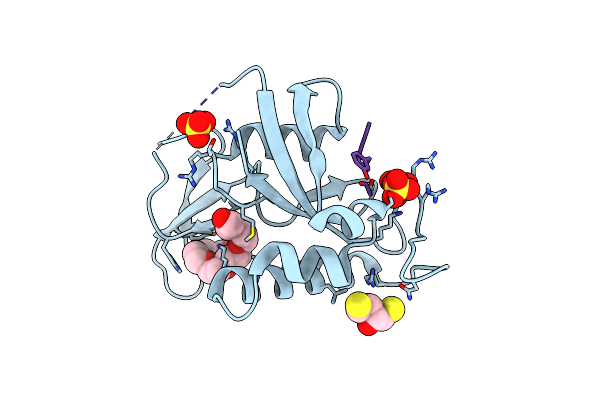
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-11-06 Classification: ISOMERASE/INHIBITOR Ligands: SO4, DTT, P33 |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-11-06 Classification: ISOMERASE/INHIBITOR Ligands: P33, DTT, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2024-11-06 Classification: ISOMERASE/INHIBITOR Ligands: A1ACH, P33, DTT, SO4 |
 |
High-Resolution Structure Of The Siderophore Periplasmic Binding Protein Ftsb From Streptococcus Pyogenes
Organism: Streptococcus pyogenes ssi-1
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2024-10-09 Classification: METAL BINDING PROTEIN Ligands: P33, PEG, EDO, P6G, ZN, NA, CL |
 |
High-Resolution Structure Of The Siderophore Periplasmic Binding Protein Ftsb Mutant Y137A From Streptococcus Pyogenes
Organism: Streptococcus pyogenes ssi-1
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2024-10-09 Classification: METAL BINDING PROTEIN Ligands: P33, PEG, ZN, SO4, CL, NA |
 |
Organism: Unidentified prokaryotic organism
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2024-09-18 Classification: HYDROLASE Ligands: EDO, P33, CA |