Search Count: 17
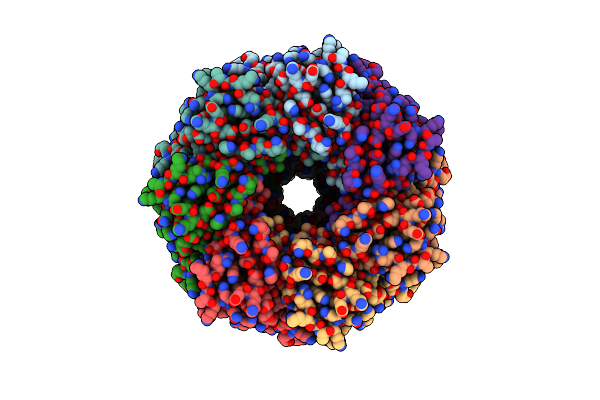 |
Alpha-Hemolysin Heptameric Pore State Bound To 10:0 Pc Lipid Chains Derived From 10:0 Pc Liposomes
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2025-06-11 Classification: LIPID BINDING PROTEIN Ligands: P1O |
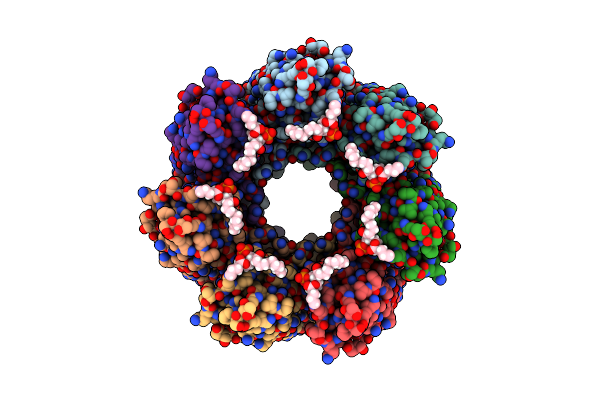 |
Alpha-Hemolysin Heptameric Pre-Pore State Bound To 10:Pc Lipid Chains Derived From 10:0 Pc Liposomes.
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2025-06-11 Classification: LIPID BINDING PROTEIN Ligands: P1O |
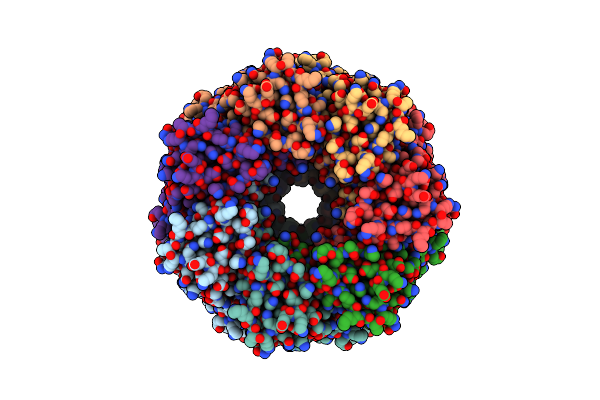 |
Alpha-Hemolysin Heptameric Late Pre-Pore State With Bound Lipids Derived From 10:0 Pc/Sphingomyelin Liposomes
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Resolution:2.60 Å Release Date: 2025-06-11 Classification: LIPID BINDING PROTEIN Ligands: P1O |
 |
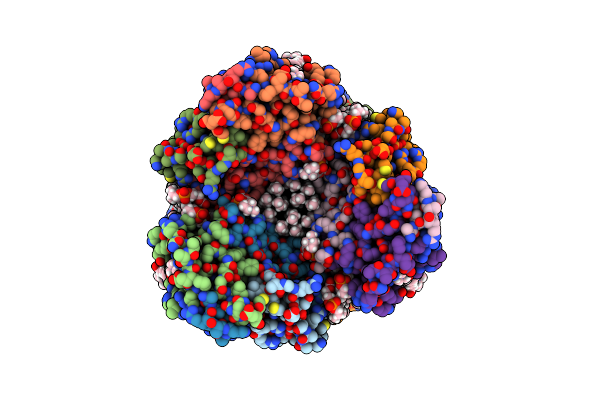
Particulate Methane Monooxygenase Crosslinked With 2,2,2-Trifluoroethanol Bound
Organism: Methylococcus capsulatus
Method: ELECTRON MICROSCOPY Release Date: 2023-11-15 Classification: OXIDOREDUCTASE Ligands: D10, CU, PLC, P1O, HXG, ETF |
 |
Particulate Methane Monooxygenase Crosslinked With 4,4,4-Trifluorobutanol Bound
Organism: Methylococcus capsulatus str. bath
Method: ELECTRON MICROSCOPY Release Date: 2023-11-15 Classification: OXIDOREDUCTASE Ligands: CU, D10, PLC, P1O, HXG, WIY |
 |
Organism: Methylococcus capsulatus str. bath
Method: ELECTRON MICROSCOPY Release Date: 2023-11-15 Classification: OXIDOREDUCTASE Ligands: CU, D10, PLC, P1O, CL, HXG |
 |
Organism: Methylococcus capsulatus str. bath
Method: ELECTRON MICROSCOPY Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: CU, D10, PLC, P1O, HXG, ETF |
 |
Cryo-Em Structure Of Ca2+-Bound Mtmem16F N562A Mutant In Digitonin Closed/Closed
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: CA, P1O |
 |
Cryoem Structure Of Methylococcus Capsulatus (Bath) Pmmo In A Native Lipid Nanodisc At 2.14 Angstrom Resolution
Organism: Methylococcus capsulatus str. bath
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: OXIDOREDUCTASE Ligands: CU, D10, PLC, HXG, P1O |
 |
Cryoem Structure Of Methylococcus Capsulatus (Bath) Pmmo In A Native Lipid Nanodisc At 2.26 Angstrom Resolution
Organism: Methylococcus capsulatus str. bath
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: OXIDOREDUCTASE Ligands: CU, D10, PLC, HXG, P1O |
 |
Cryoem Structure Of Methylococcus Capsulatus (Bath) Pmmo In A Native Lipid Nanodisc At 2.16 Angstrom Resolution
Organism: Methylococcus capsulatus str. bath
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: OXIDOREDUCTASE Ligands: CU, D10, PLC, HXG, P1O |
 |
Cryoem Structure Of Methylococcus Capsulatus (Bath) Pmmo In A Native Lipid Nanodisc At 2.34 Angstrom Resolution
Organism: Methylococcus capsulatus str. bath
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: OXIDOREDUCTASE Ligands: CU, D10, PLC, HXG, P1O |
 |
Cryoem Structure Of Methylococcus Capsulatus (Bath) Pmmo Treated With Potassium Cyanide In A Native Lipid Nanodisc At 3.65 Angstrom Resolution
Organism: Methylococcus capsulatus str. bath
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: OXIDOREDUCTASE Ligands: CU, P1O, PLC, D10 |
 |
Cryoem Structure Of Methylococcus Capsulatus (Bath) Pmmo Treated With Potassium Cyanide And Copper In A Native Lipid Nanodisc At 3.62 Angstrom Resolution
Organism: Methylococcus capsulatus str. bath
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: OXIDOREDUCTASE Ligands: CU, D10, PLC, HXG, P1O |
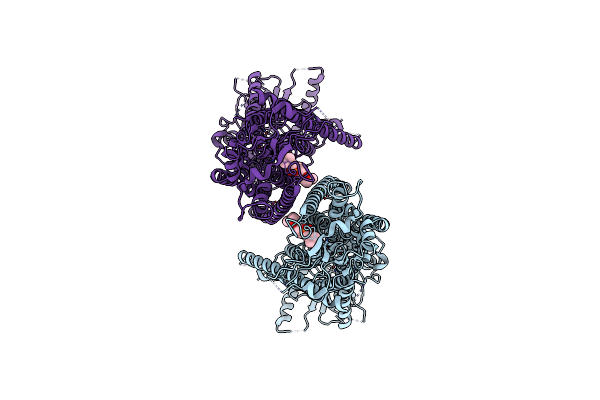 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2019-03-06 Classification: MEMBRANE PROTEIN Ligands: CA, P1O |
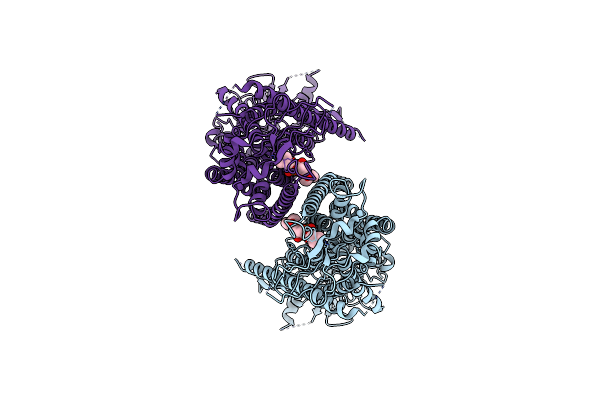 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2019-03-06 Classification: MEMBRANE PROTEIN Ligands: P1O |
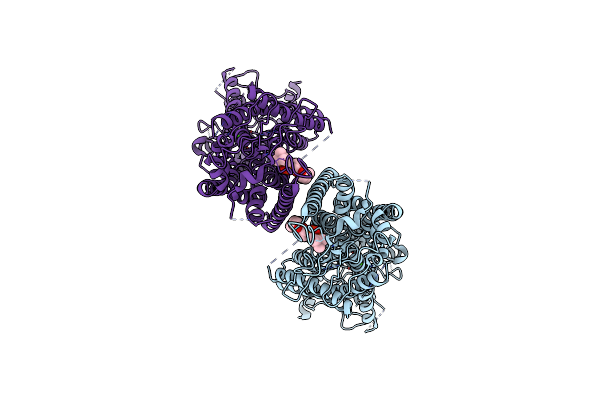 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2019-03-06 Classification: MEMBRANE PROTEIN Ligands: CA, P1O |

