Search Count: 16
 |
Crystal Structure Of Trehalose Synthase Mutant N253C From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-03-19 Classification: ISOMERASE Ligands: CA, MG, TRS |
 |
Crystal Structure Of Trehalose Synthase Mutant N253E From Deinococcus Radiodurans
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / ccug 27074 / lmg 4051 / nbrc 15346 / ncimb 9279 / vkm b-1422 / r1)
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: CA, MG, TRS |
 |
Crystal Structure Of Trehalose Synthase Mutant N253Q From Deinococcus Radiodurans
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / ccug 27074 / lmg 4051 / nbrc 15346 / ncimb 9279 / vkm b-1422 / r1)
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: CA, MG, TRS |
 |
Crystal Structure Of Trehalose Synthase Mutant N253T From Deinococcus Radiodurans
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / ccug 27074 / lmg 4051 / nbrc 15346 / ncimb 9279 / vkm b-1422 / r1)
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: CA, MG, TRS |
 |
Crystal Structure Of Trehalose Synthase Mutant R148A From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: CA, MG, TRS |
 |
Crystal Structure Of Trehalose Synthase From Deinococcus Radiodurans Complexed With Validoxylamine A (Vaa)
Organism: Deinococcus radiodurans r1 = atcc 13939 = dsm 20539
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: CA, MG, VDM |
 |
Crystal Structure Of Trehalose Synthase Mutamt E324D From Deinococcus Radiodurans Complexed With Validoxylamine A (Vaa)
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / ccug 27074 / lmg 4051 / nbrc 15346 / ncimb 9279 / vkm b-1422 / r1)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: CA, MG, VDM |
 |
Crystal Structure Of Trehalose Synthase Mutant N253H From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: CA, MG, TRS |
 |
Cryo-Em Structure Of Coagulation Factor Beta-Xiia In Complex With The Garadacimab Fab Fragment (Symmetric Dimer)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-12-13 Classification: BLOOD CLOTTING |
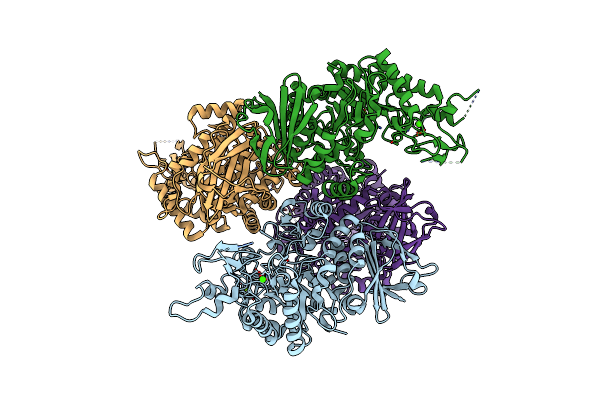 |
The N253F Mutant Structure Of Trehalose Synthase From Deinococcus Radiodurans Reveals An Open Active-Site Conformation
Organism: Deinococcus radiodurans str. r1
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2017-10-25 Classification: ISOMERASE Ligands: CA, MG |
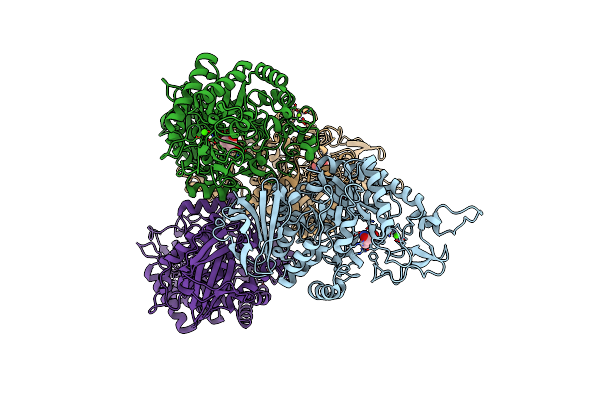 |
The N253R Mutant Structures Of Trehalose Synthase From Deinococcus Radiodurans Display Two Different Active-Site Conformations
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:2.93 Å Release Date: 2017-08-30 Classification: ISOMERASE Ligands: CA, MG, TRS |
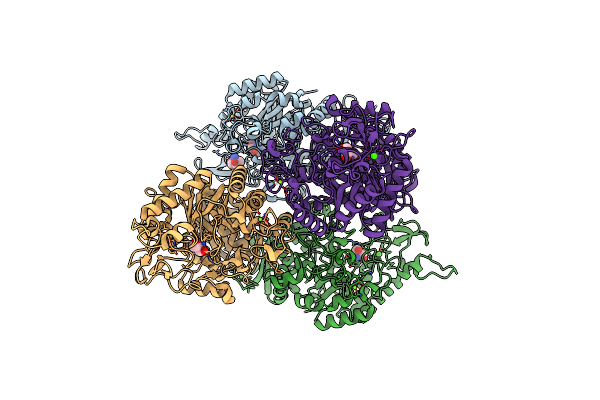 |
Crystal Structures Of Trehalose Synthase From Deinococcus Radiodurans Reveal That A Closed Conformation Is Involved In The Intramolecular Isomerization Catalysis
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2014-12-24 Classification: ISOMERASE Ligands: CA, MG, TRS |
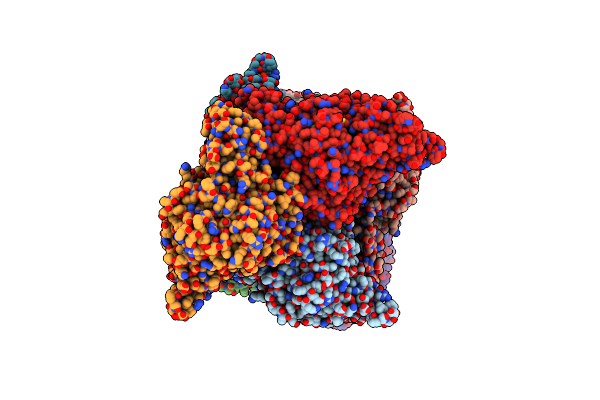 |
Crystal Structure Of Trehalose Synthase From Deinococcus Radiodurans Reveals A Closed Conformation For Catalysis Of The Intramolecular Isomerization
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-12-17 Classification: ISOMERASE Ligands: CA, MG, TRS |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-01-25 Classification: LIGASE Ligands: ZN |
 |
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2010-01-26 Classification: HYDROLASE |
 |
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:3.92 Å Release Date: 2010-01-26 Classification: HYDROLASE |

