Search Count: 19
 |
Organism: Pseudomonas sp. uk4
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: PROTEIN FIBRIL |
 |
Organism: Rhodonellum psychrophilum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-05-14 Classification: ISOMERASE |
 |
Organism: Rhodococcus sp. jg-3
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2025-05-14 Classification: ISOMERASE Ligands: TRS, NA |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2025-02-05 Classification: HYDROLASE Ligands: PG6, SO4 |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2025-02-05 Classification: HYDROLASE Ligands: SO4, PMS |
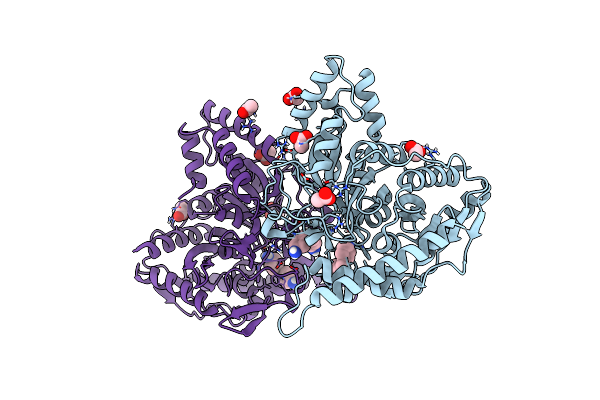 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-02-05 Classification: HYDROLASE Ligands: ACT, A1IHK |
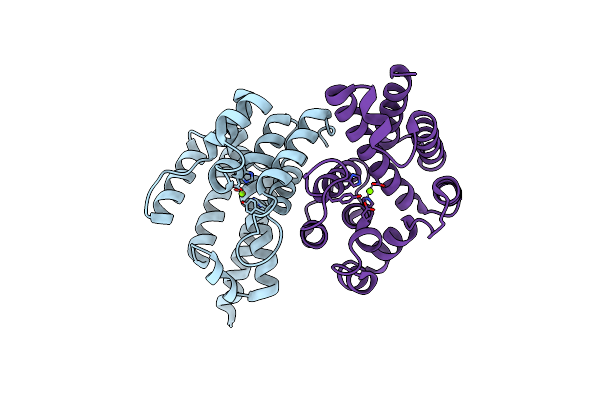 |
Se-Met Derivative Structure Of A Small Alarmone Hydrolase (Relh) From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: MG |
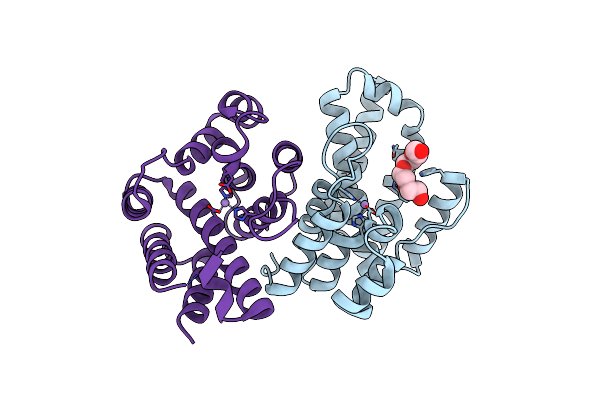 |
Native Structure Of A Small Alarmone Hydrolase (Relh) From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: MN, PG4 |
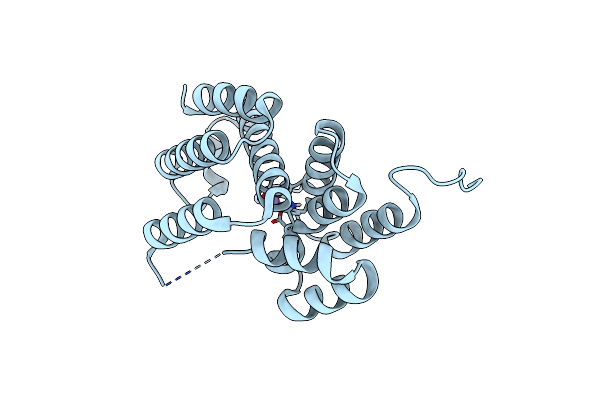 |
Organism: Leptospira levettii
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: MN |
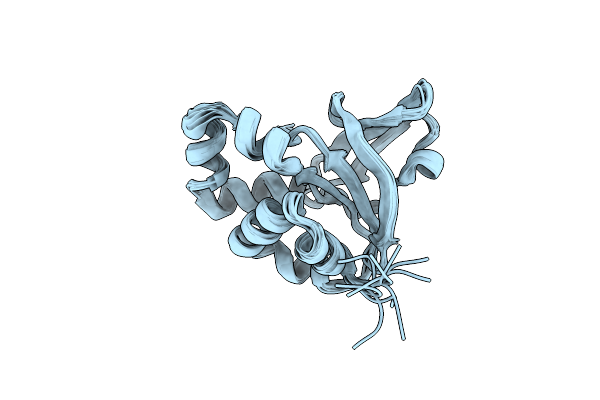 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2013-08-21 Classification: UNKNOWN FUNCTION |
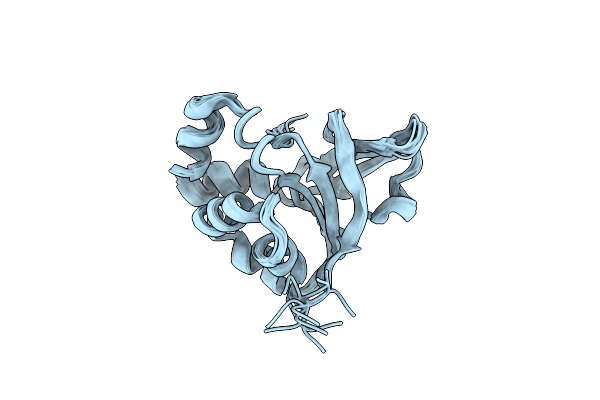 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2013-08-21 Classification: UNKNOWN FUNCTION |
 |
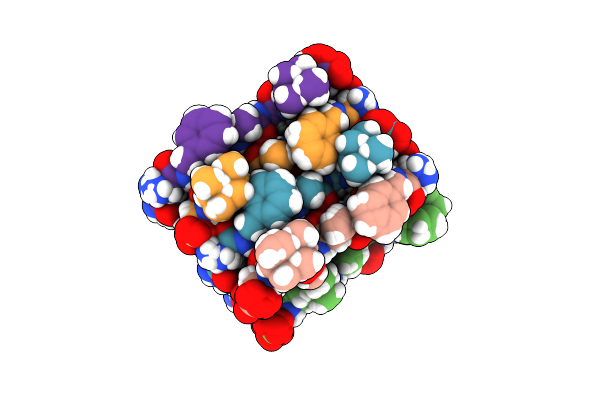
Organism: Eurotium amstelodami
Method: SOLUTION NMR Release Date: 2012-10-31 Classification: ANTIMICROBIAL PROTEIN |
 |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2005-10-26 Classification: RIBOSOMAL PROTEIN |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-10-26 Classification: RIBOSOMAL PROTEIN |
 |
Protein Aggregation And Alzheimer'S Disease: Crystallographic Analysis Of The Phenomenon. Engineered Version Of The Ribosomal Protein S6 Used As A Stable Scaffold To Study Oligomerization.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2000-09-08 Classification: RIBOSOMAL PROTEIN |
 |
Protein Aggregation And Alzheimer'S Disease: Crystallographic Analysis Of The Phenomenon. Engineered Version Of The Ribosomal Protein S6 Used As A Stable Scaffold To Study Oligomerization.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2000-09-08 Classification: RIBOSOMAL PROTEIN |
 |
Protein Aggregation And Alzheimer'S Disease: Crystallographic Analysis Of The Phenomenon. Engineered Version Of The Ribosomal Protein S6 Used As A Stable Scaffold To Study Oligomerization.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2000-06-29 Classification: RIBOSOMAL PROTEIN Ligands: MG |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 1998-11-30 Classification: RIBOSOME |

