Search Count: 30
 |
Characterization Of The Sars-Cov-2 S Protein: Biophysical, Biochemical, Structural, And Antigenic Analysis
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-06-10 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Listeria phage a500
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2019-10-30 Classification: VIRAL PROTEIN |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-03-30 Classification: VIRAL PROTEIN Ligands: 5VE |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-03-30 Classification: VIRAL PROTEIN Ligands: NAG, 5VG |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-03-30 Classification: VIRAL PROTEIN Ligands: FMT, NAG, 5VF |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2016-03-30 Classification: VIRAL PROTEIN Ligands: NAG, 5VH |
 |
Structure Of Cbm32-3 From A Family 31 Glycoside Hydrolase From Clostridium Perfringens In Complex With N-Acetylgalactosamine
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-10-07 Classification: SUGAR BINDING PROTEIN Ligands: NGA, CA |
 |
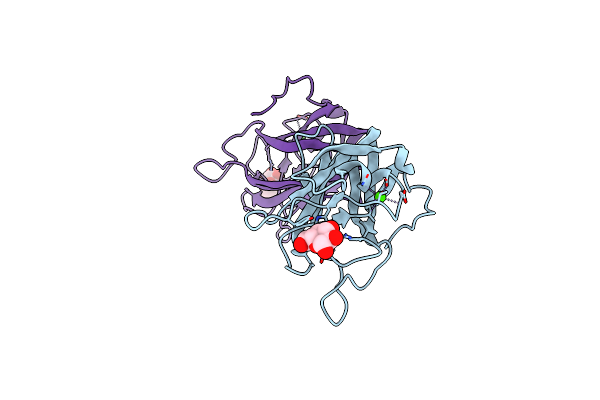
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-10-07 Classification: SUGAR BINDING PROTEIN Ligands: NGA, CA, GOL |
 |
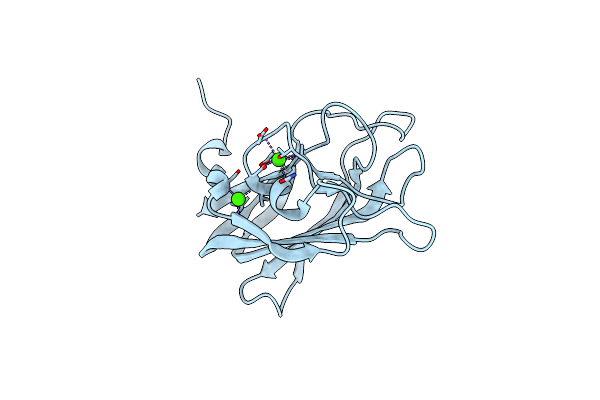
Structure Of Cbm32-3 From A Family 31 Glycoside Hydrolase From Clostridium Perfringens In Complex With Galactose
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-12-24 Classification: SUGAR BINDING PROTEIN Ligands: CA, GAL |
 |
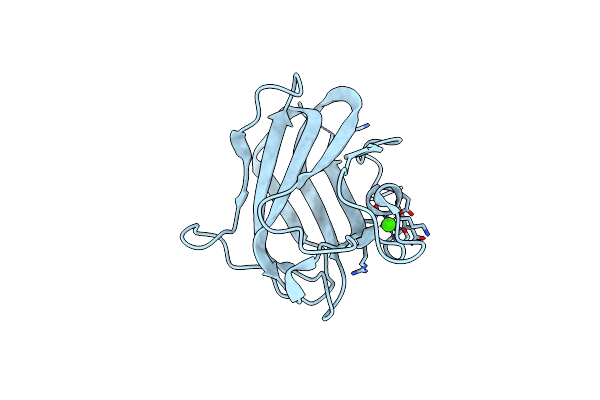
Structure Of Cbm32-3 From A Family 31 Glycoside Hydrolase From Clostridium Perfringens
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2014-07-23 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
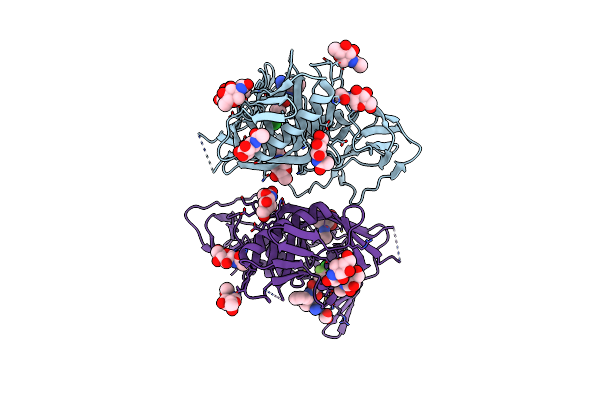
Structure Of Cbm32-1 From A Family 31 Glycoside Hydrolase From Clostridium Perfringens
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2014-07-16 Classification: SUGAR BINDING PROTEIN Ligands: CA, MG |
 |
Crystal Structure Of Clade C1086 Hiv-1 Gp120 Core In Complex With Dmj-Ii-121
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-05-29 Classification: VIRAL PROTEIN/INHIBITOR Ligands: 1C1, NAG, FMT |
 |
Crystal Structure Of Clade A/E 93Th057 Hiv-1 Gp120 H375S Core In Complex With Dmj-Ii-121
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-05-29 Classification: VIRAL PROTEIN/INHIBITOR Ligands: 1C1, EPE, NAG |
 |
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2013-05-01 Classification: TRANSFERASE Ligands: PO4, GOL |
 |
Crystal Structure Of Clade A/E 93Th057 Hiv-1 Gp120 Core In Complex With Ts-Ii-224
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2012-05-02 Classification: VIRAL PROTEIN/INHIBITOR Ligands: NAG, EPE, 0LM |
 |
Crystal Structure Of Clade A/E 93Th057 Hiv-1 Gp120 Core In Complex With Aws-I-50
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-05-02 Classification: VIRAL PROTEIN/INHIBITOR Ligands: NAG, EPE, 0LL |
 |
Crystal Structure Of Clade A/E 93Th057 Hiv-1 Gp120 Core In Complex With Dmj-I-228
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2012-05-02 Classification: VIRAL PROTEIN/INHIBITOR Ligands: NAG, EPE, 0LK |
 |
Crystal Structure Of Clade A/E 93Th057 Hiv-1 Gp120 Core In Complex With Aws-I-169
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-05-02 Classification: VIRAL PROTEIN/INHIBITOR Ligands: NAG, EPE, 0LJ |
 |
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-10-13 Classification: TRANSFERASE Ligands: SO4, NHE |
 |
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-10-13 Classification: TRANSFERASE |

