Search Count: 51
 |
Crystal Structure Of Maltose Binding Protein (Apo), Mutant Trp10 To 4-Cyanotryptophan
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2024-12-18 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2024-12-18 Classification: SUGAR BINDING PROTEIN Ligands: PEG, EDO, PGE, NA, CD |
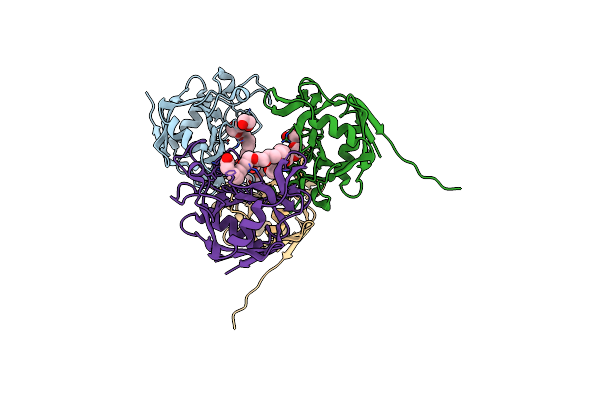 |
E. Coli Peptidyl-Prolyl Cis-Trans Isomerase Containing (2S,3S)-4-Fluorovaline
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2024-10-09 Classification: ISOMERASE Ligands: 1PE, EDO, PGE, ACT, PEG, XPE |
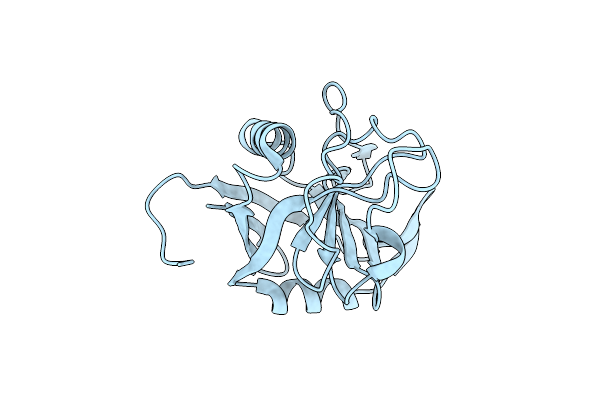 |
E. Coli Peptidyl-Prolyl Cis-Trans Isomerase Containing Delta1-Monofluoro-Leucines
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2024-05-22 Classification: ISOMERASE |
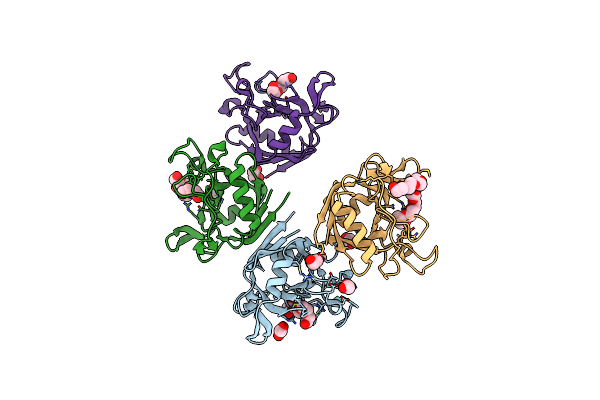 |
E. Coli Peptidyl-Prolyl Cis-Trans Isomerase Containing Delta2-Monofluoro-Leucines
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-05-22 Classification: ISOMERASE Ligands: EDO, PGE, PEG, 1PE, XPE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-05-22 Classification: ISOMERASE Ligands: PEG, PGE, EDO, PG4, 2PE |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-03-16 Classification: Hydrolase/Hydrolase Inhibitor |
 |
Organism: Methanogenic archaeon mixed culture iso4-g1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-03-02 Classification: TRANSLATION Ligands: EDO, SO4 |
 |
E. Coli Peptidyl-Prolyl Cis-Trans Isomerase, Mutant Phe27Cf3-Tyr/Phe98Cf3-Tyr
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-09-29 Classification: ISOMERASE |
 |
E. Coli Peptidyl-Prolyl Cis-Trans Isomerase, Mutant Phe4Ala Phe27Cf3-Phe/Phe98Cf3-Phe
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2021-09-29 Classification: ISOMERASE |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2020-05-13 Classification: LYASE Ligands: CL, EPE |
 |
Endoplasmic Reticulum Protein 29 (Erp29) C-Terminal Domain: Structure Determination From Backbone Amide Pseudocontact Shifts Generated By Double-Histidine Cobalt Tags
|
 |
Organism: Zika virus, Zika virus (strain mr 766), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-06-26 Classification: VIRAL PROTEIN |
 |
1.8 Angstrom Crystal Structure Of Imp-1 Metallo-Beta-Lactamase With A Mixed Iron-Zinc Center In The Active Site
Organism: Pseudomonas aeruginosa ncgm2.s1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-09-17 Classification: HYDROLASE Ligands: FE, ZN, FLC |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-12-18 Classification: DNA BINDING PROTEIN |
 |
Endoplasmic Reticulum Protein 29 (Erp29) C-Terminal Domain: 3D Protein Fold Determination From Backbone Amide Pseudocontact Shifts Generated By Lanthanide Tags At Multiple Sites
|
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-04-03 Classification: TRANSFERASE Ligands: CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2013-04-03 Classification: TRANSFERASE |
 |
Solution Structure Of The R3H Domain From Human Smubp-2 In Complex With 2'-Deoxyguanosine-5'-Monophosphate
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2012-10-24 Classification: HYDROLASE Ligands: DGP |
 |
Organism: Bacillus subtilis
Method: SOLUTION NMR Release Date: 2009-03-03 Classification: REPLICATION Ligands: ZN |

