Search Count: 426
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN |
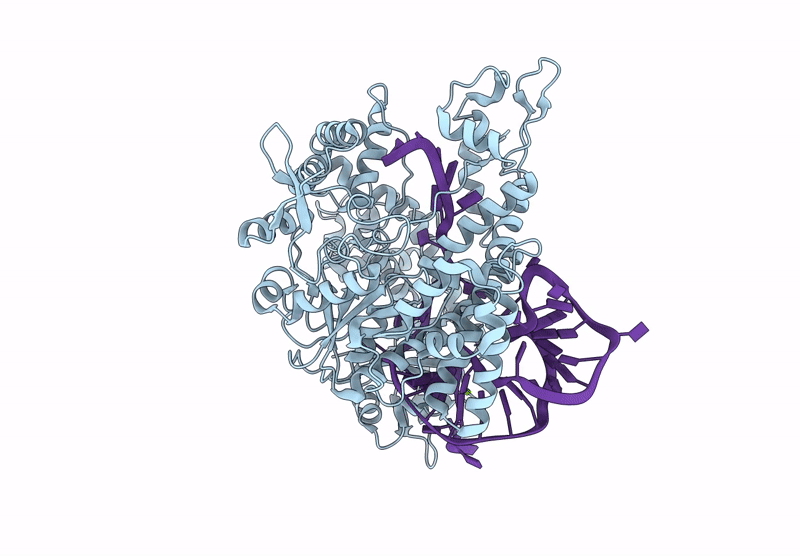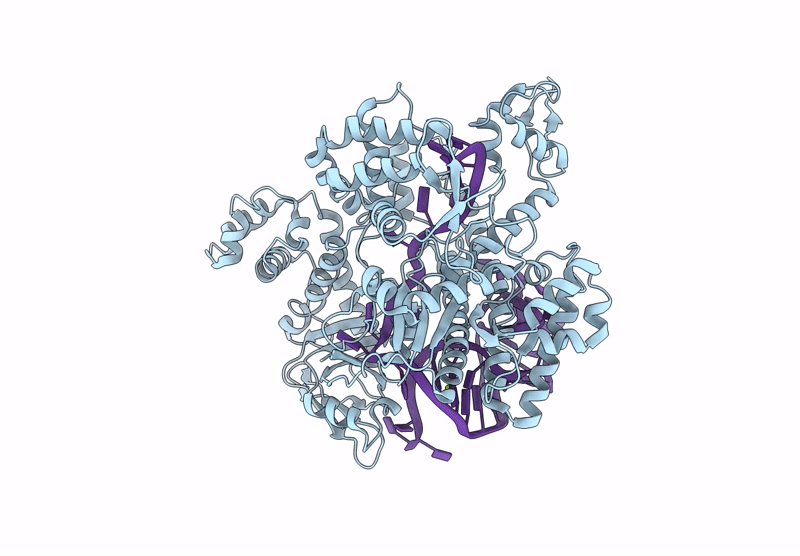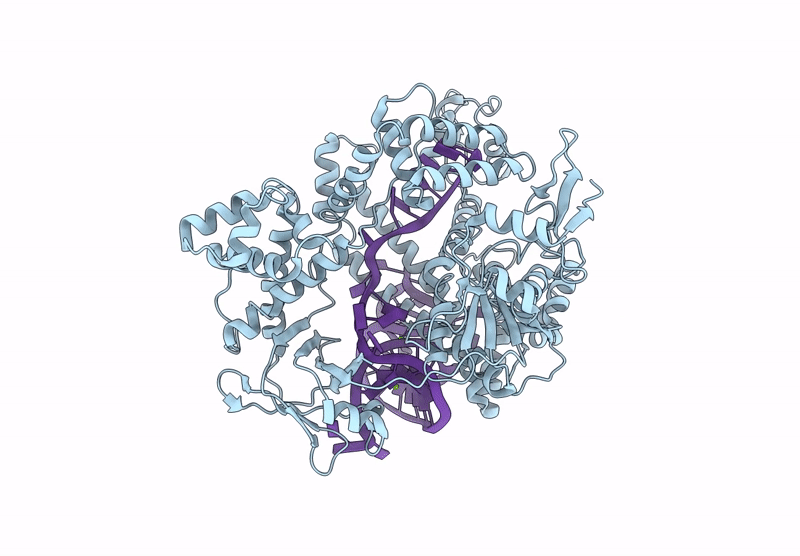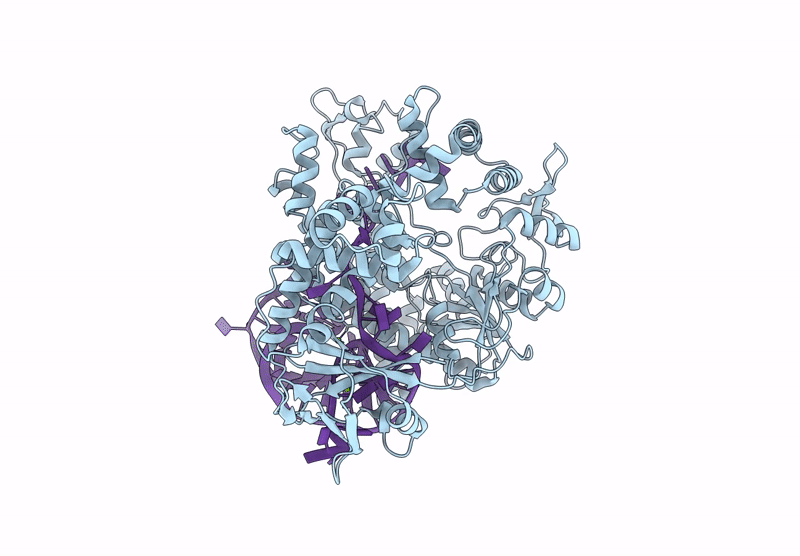 |
Organism: Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
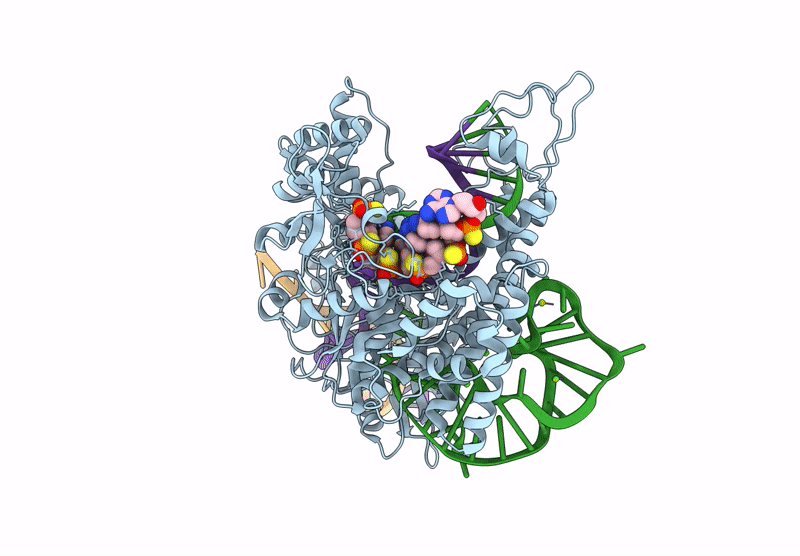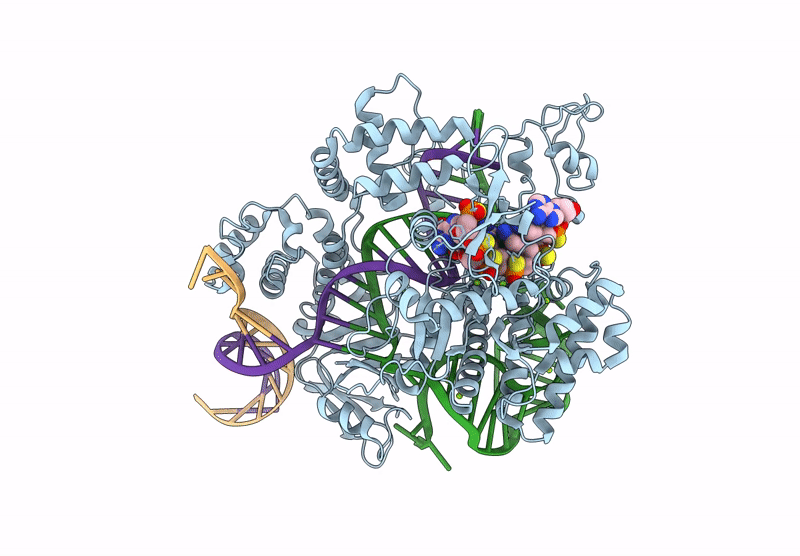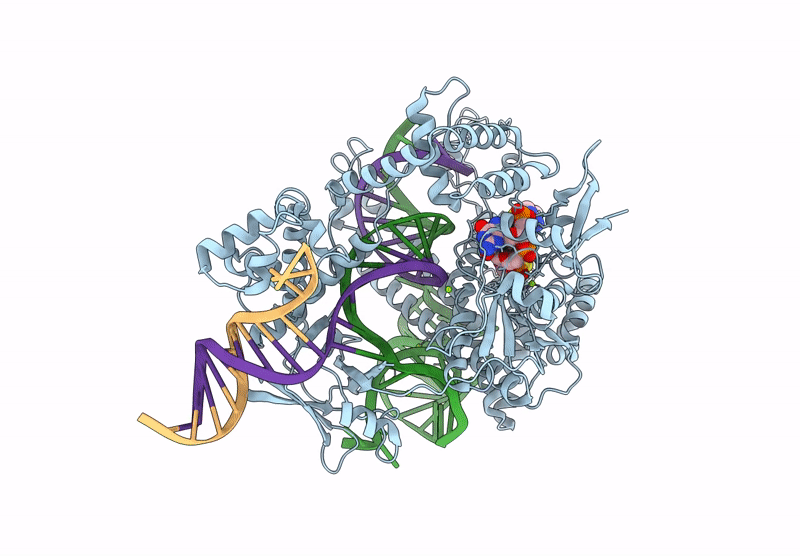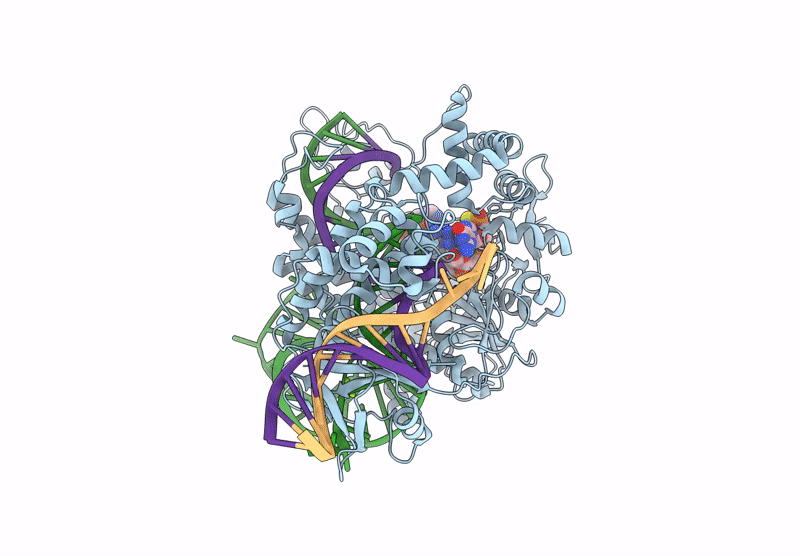
Cryo-Em Structure Of Caslambda2-Crrna-Target Dna Ternary Complex In The Incompetent State
Organism: Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryo-Em Structure Of Caslambda2-Crrna-Target Dna Ternary Complex In The Intermediate State
Organism: Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryo-Em Structure Of Caslambda2-Crrna-Target Dna Ternary Complex In The Nts-Cleaving State
Organism: Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: MG, GS, SC, AS |
 |
Cryo-Em Structure Of Caslambda2-Crrna-Target Dna Ternary Complex In The Ts-Cleaving State
Organism: Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Organism: Bacteroides caccae
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: LYASE Ligands: MG |
 |
Organism: Bacteroides caccae
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: LYASE |
 |
Structure Of An Inactive Beta-D-Glucuronate Dehydratase Mutant In Complex With Chondrosine
Organism: Bacteroides caccae
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: LYASE Ligands: IOD |
 |
Crystal Structure Of A Parallel Eight-Helix Coiled Coil Cc-Type2-Ii-I17V-I21F
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-02-26 Classification: DE NOVO PROTEIN |
 |
O-Glcnacase (Oga) Inhibitor Complex For The Treatment Of Alzheimer'S Disease
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: A1AKM |
 |
O-Glcnacase (Oga) Inhibitor Complex For The Treatment Of Alzheimer'S Disease
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: A1AKL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-11-20 Classification: LIGASE Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: LIGASE Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-11-20 Classification: LIGASE Ligands: MPD |
 |
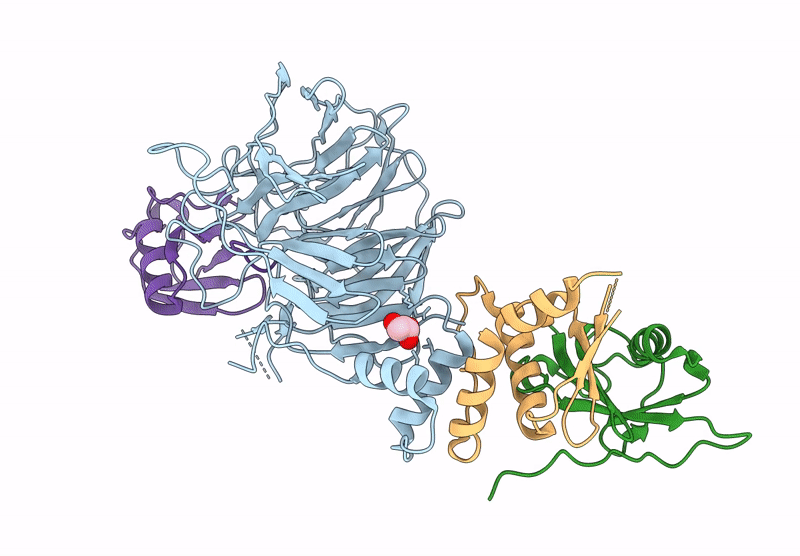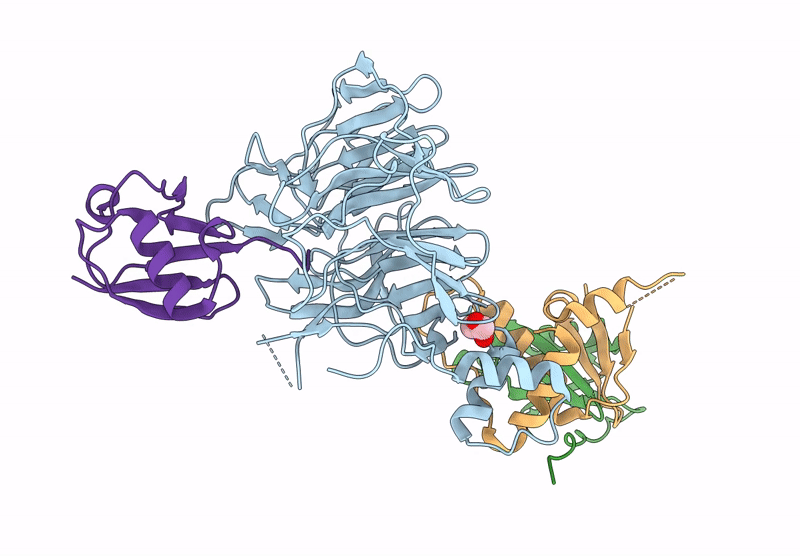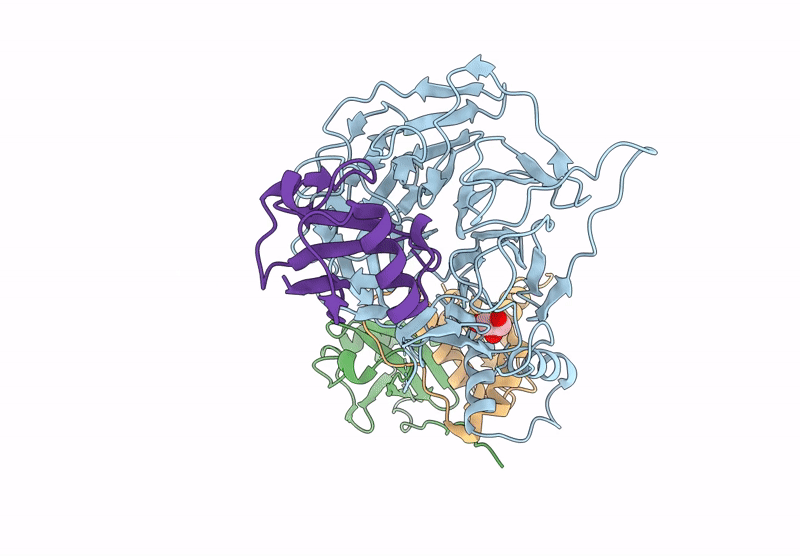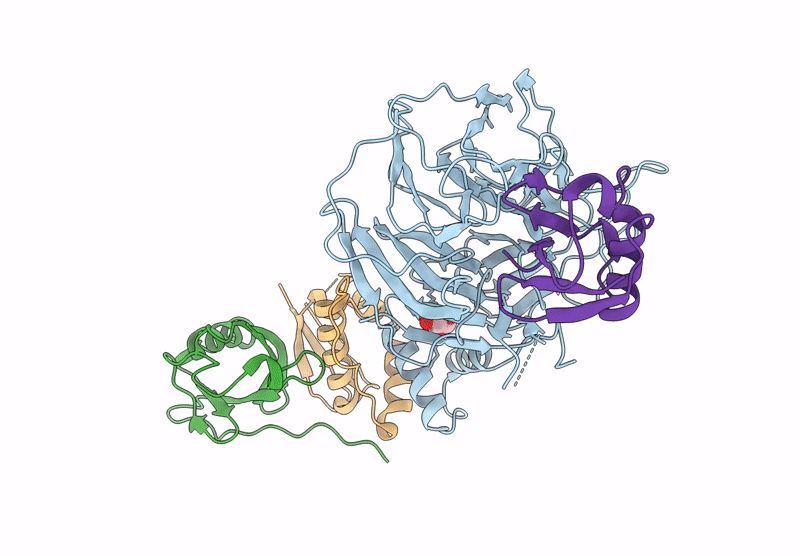
Focused Map Of Cryo-Em Structure Of Ubiquitin C-Degron Bound To Klhdc10-Elob/C
|
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2024-11-06 Classification: LIGASE/INHIBITOR Ligands: A1APK, NCO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-11-06 Classification: LIGASE/INHIBITOR Ligands: A1APS, NCO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-11-06 Classification: LIGASE/INHIBITOR Ligands: A1APT, NCO |
 |
Organism: Clostridium perfringens atcc 13124
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-09-04 Classification: HYDROLASE Ligands: NO3, NA, EDO, 5N6 |

