Search Count: 33
 |
Organism: Bacillus subtilis
Method: SOLUTION NMR, SOLUTION SCATTERING Release Date: 2025-06-25 Classification: GENE REGULATION Ligands: ZN |
 |
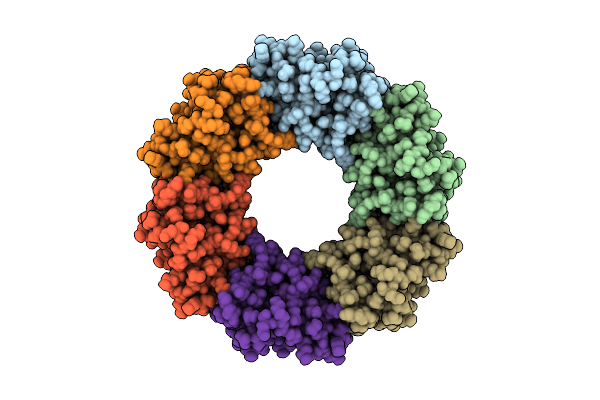
Alkalihalobacillus Halodurans (Aha) Trp Rna Binding Attenuation Protein (Trap) Mutant Dtrap With Trp
Organism: Halalkalibacterium halodurans
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: RNA BINDING PROTEIN Ligands: TRP |
 |
Alkalihalobacillus Halodurans (Aha) Trp Rna Binding Attenuation Protein (Trap) Mutant Dtrap Without Trp
Organism: Halalkalibacterium halodurans
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: RNA BINDING PROTEIN |
 |
Alkalihalobacillus Halodurans (Aha) Trp Rna Binding Attenuation Protein (Trap) Mutant T49A/T52A Dtrap With Trp
Organism: Halalkalibacterium halodurans
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: RNA BINDING PROTEIN Ligands: TRP |
 |
Organism: Caulobacter vibrioides
Method: SOLUTION NMR Release Date: 2024-10-16 Classification: RNA BINDING PROTEIN |
 |
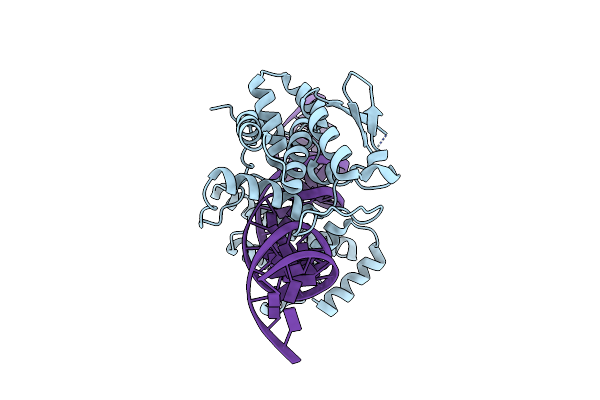
Organism: Escherichia phage p1
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: RECOMBINATION/DNA |
 |
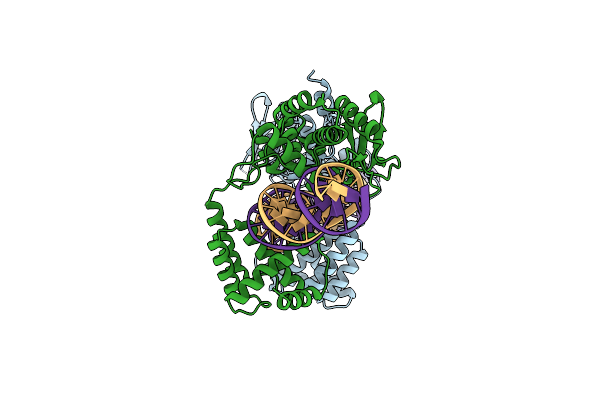
Organism: Escherichia phage p1
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: RECOMBINATION/DNA |
 |
Heterodimer Of Cre Recombinase Mutants D33A/A36V/R192A And R72E/L115D/R119D In Complex With Loxp Dna.
Organism: Escherichia phage p1
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: RECOMBINATION/DNA |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Resolution:15.00 Å Release Date: 2020-07-29 Classification: EXOCYTOSIS |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-05-09 Classification: LYASE Ligands: MG, CO3 |
 |
Organism: Pseudomonas sp. hmsc75e02
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-05-09 Classification: LYASE Ligands: ACY, EDO, NI |
 |
Organism: Pseudomonas sp. hmsc75e02
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-05-09 Classification: LYASE Ligands: CA, ACY, EDO |
 |
Organism: Caulobacter crescentus (strain atcc 19089 / cb15)
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2017-08-09 Classification: RNA BINDING PROTEIN |
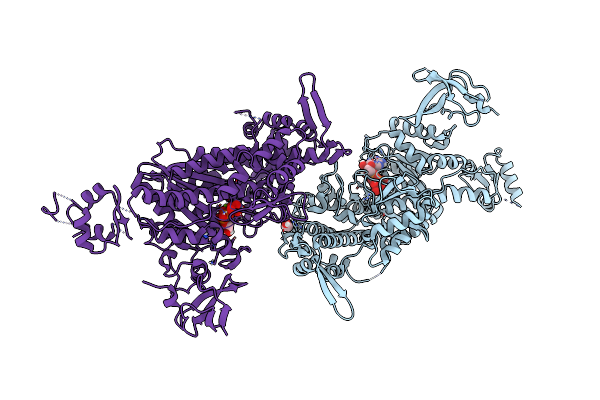 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2016-03-23 Classification: MOTOR PROTEIN Ligands: MG, VO4, ADP, EDO |
 |
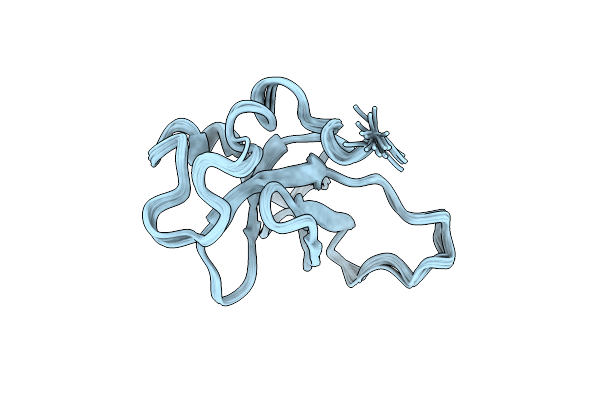
Organism: Homo sapiens, Moloney murine leukemia virus
Method: SOLUTION NMR Release Date: 2016-03-02 Classification: PROTEIN BINDING |
 |
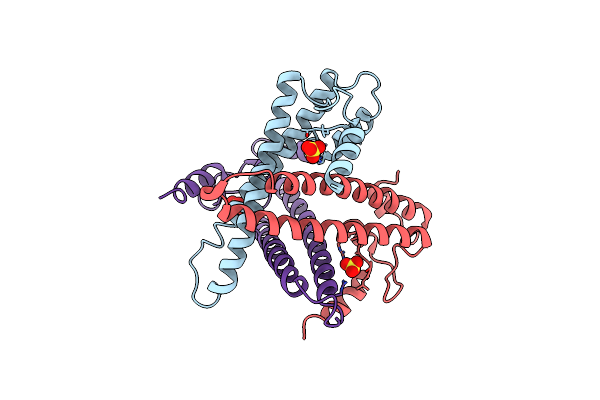 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2013-02-27 Classification: HYDROLASE Ligands: SO4 |
 |
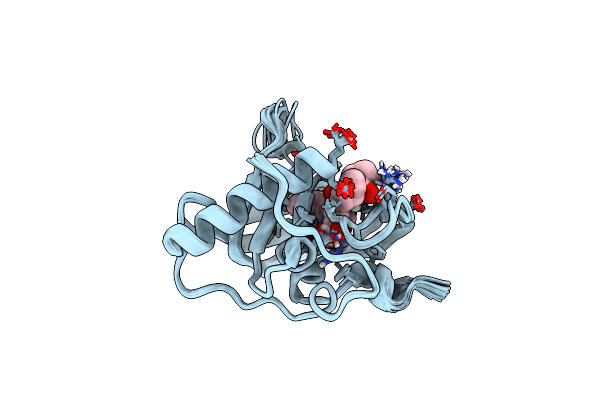
Organism: Pyrococcus furiosus dsm 3638
Method: SOLUTION NMR Release Date: 2009-09-15 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Escherichia coli k-12
Method: SOLUTION NMR Release Date: 2009-08-25 Classification: HYDROLASE/ANTIBIOTIC Ligands: ZN, BB2 |
 |
Organism: Pyrococcus furiosus
Method: SOLUTION NMR Release Date: 2008-12-02 Classification: HYDROLASE Ligands: ZN |

