Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
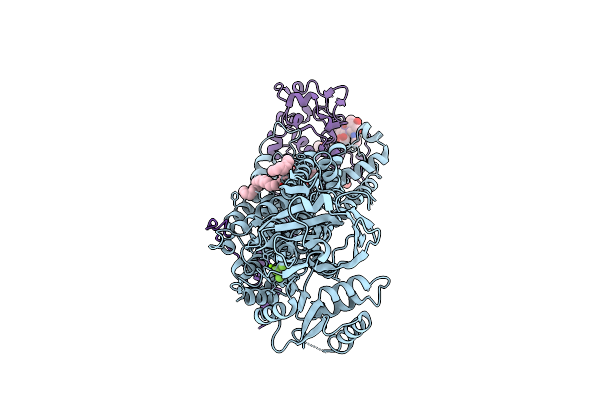 |
Cryo-Em Structure Of A Human Flippase Mutant Atp11C Q79E-Cdc50A In Ptdcho-Occluded E2-Alf State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: ALF, MG, PCW, NAG |
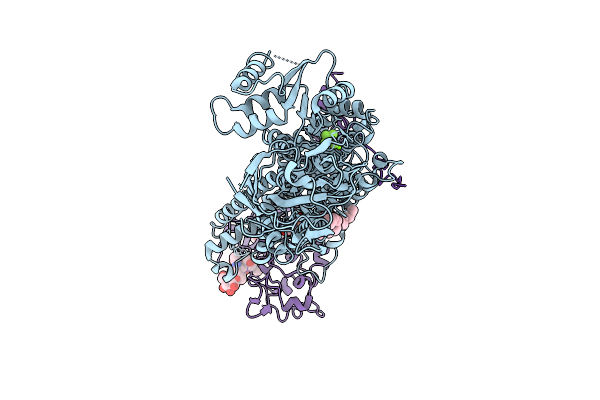 |
Cryo-Em Structure Of A Human Flippase Mutant Atp11C Q79E-Cdc50A In Ptdser-Occluded E2P-Alf State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: ALF, MG, P5S, NAG |
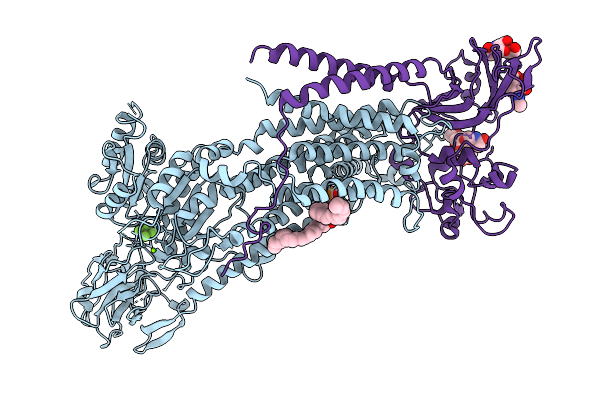 |
Cryo-Em Structure Of A Human Flippase Mutant Atp11C Q79A-Cdc50A In The Ptdser-Occluded E2-Alf State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: ALF, MG, P5S, NAG |
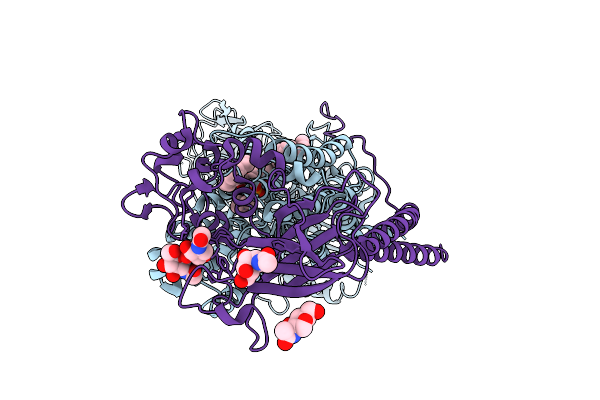 |
Cryo-Em Structrure Of A Human Flippase Mutant Atp11C Q79E-Cdc50A In Ptdcho-Open E2-Bef State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: MG, PCW, NAG |
 |
Organism: Escherichia coli, Homo sapiens, Synthetic construct, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: MEMBRANE PROTEIN Ligands: NKP |
 |
Organism: Homo sapiens, Mus musculus, Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: MEMBRANE PROTEIN Ligands: NKP |
 |
Organism: Escherichia coli, Homo sapiens, Synthetic construct, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: MEMBRANE PROTEIN Ligands: NKP |
 |
Organism: Escherichia coli, Homo sapiens, Synthetic construct, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: SIGNALING PROTEIN Ligands: NAG, OKL |
 |
Organism: Homo sapiens, Synthetic construct, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: SIGNALING PROTEIN Ligands: NAG, FI7 |
 |
Organism: Escherichia coli, Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: SIGNALING PROTEIN Ligands: NAG, P8A |
 |
Organism: Homo sapiens, Synthetic construct, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: SIGNALING PROTEIN Ligands: NAG, P9X |
 |
Organism: Escherichia coli, Homo sapiens, Synthetic construct, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: SIGNALING PROTEIN Ligands: P9X |
 |
Organism: Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: SIGNALING PROTEIN Ligands: P9X |
 |
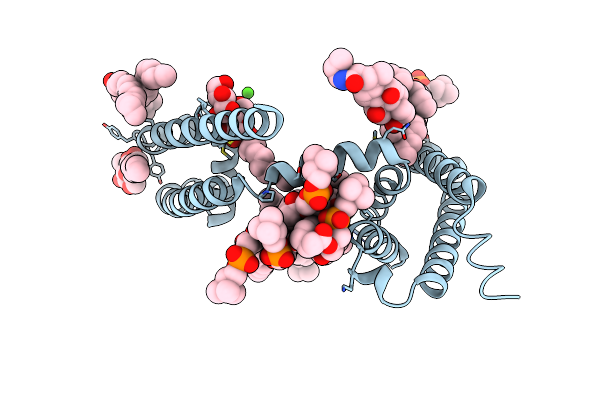
Crystal Structure Of Voltage-Gated Sodium Channel Navab N49K Mutant In Sodium Ion Condition
Organism: Aliarcobacter butzleri
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2023-07-26 Classification: MEMBRANE PROTEIN Ligands: LMT, 1N7, PX4, NA |
 |
Crystal Structure Of Voltage-Gated Sodium Channel Navab N49K Mutant In Calcium Ion Condition
Organism: Aliarcobacter butzleri
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-07-26 Classification: MEMBRANE PROTEIN Ligands: LMT, 1N7, PX4, CA |
 |
Crystal Structure Of Voltage-Gated Sodium Channel Navab N49K/L176Q Mutant In Sodium Ion Condition
Organism: Aliarcobacter butzleri
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2023-07-26 Classification: MEMBRANE PROTEIN Ligands: LMT, 1N7, PX4, NA |
 |
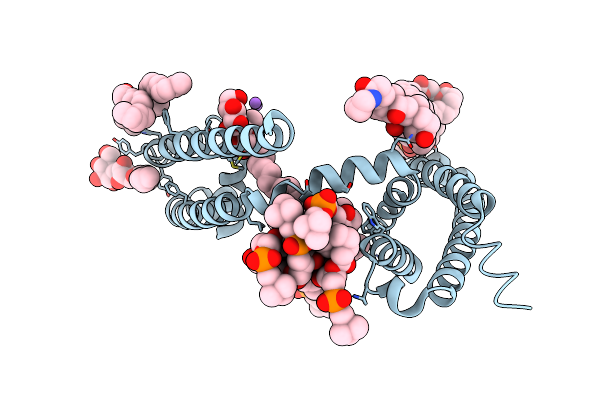
Crystal Structure Of Voltage-Gated Sodium Channel Navab N49K/L176Q Mutant In Calcium Ion Condition
Organism: Aliarcobacter butzleri
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2023-07-26 Classification: MEMBRANE PROTEIN Ligands: LMT, 1N7, PX4, CA |
 |
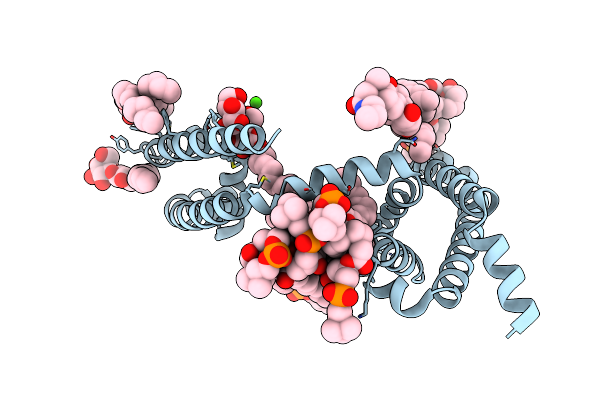
Crystal Structure Of Voltage-Gated Sodium Channel Navab N49K/L176G Mutant In Sodium Ion Condition
Organism: Aliarcobacter butzleri
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2023-07-26 Classification: MEMBRANE PROTEIN Ligands: LMT, 1N7, PX4, NA |
 |
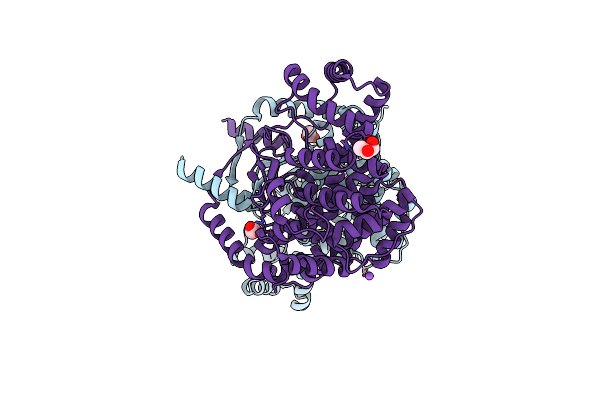
Crystal Structure Of Voltage-Gated Sodium Channel Navab N49K/L176G Mutant In Calcium Ion Condition
Organism: Aliarcobacter butzleri
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2023-07-26 Classification: MEMBRANE PROTEIN Ligands: LMT, 1N7, PX4, CA |
 |
Crystal Structure Of Mitochondrial Citrate Synthase (Cit1) From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2023-04-26 Classification: TRANSFERASE |