Search Count: 17
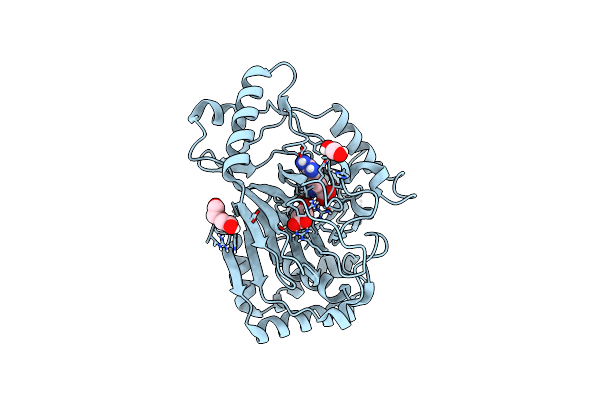 |
Crystal Structure Of The L-Arginine Hydroxylase Vioc Mehis316, Bound To Fe(Ii), L-Arginine, And Succinate
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: OXIDOREDUCTASE Ligands: FE2, SIN, ARG, EDO, PEG |
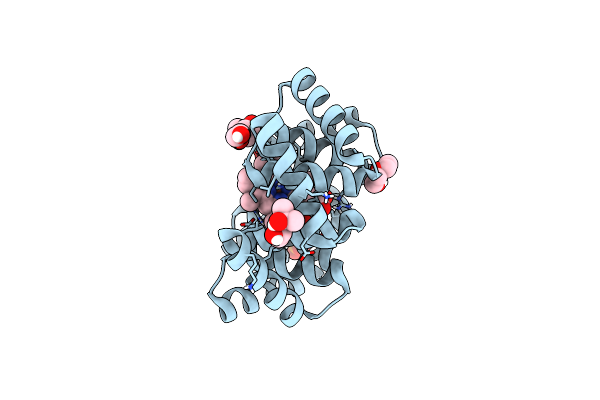 |
Crystal Structure Of A Computationally Designed Heme Binding Protein, Dnhem1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-07-05 Classification: BIOSYNTHETIC PROTEIN Ligands: IMD, HEM, MPD, PO4, PEG, EDO |
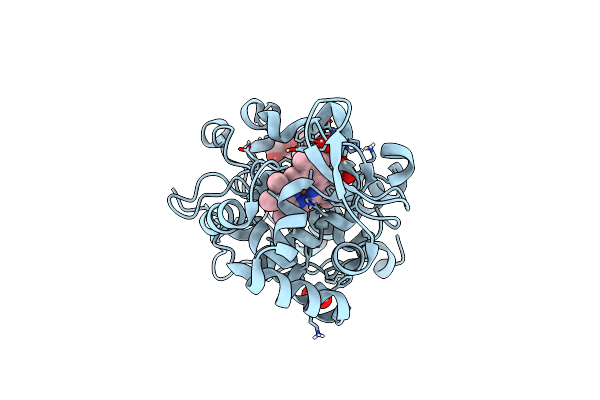 |
The Crystal Structure Of Engineered Cytochrome C Peroxidase From Saccharomyces Cerevisiae With A Trp51 To S-Trp51 Modification
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-06-16 Classification: OXIDOREDUCTASE Ligands: HEM, EDO, 1PE |
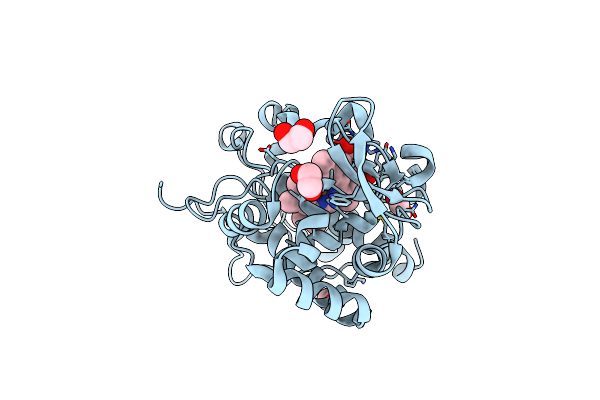 |
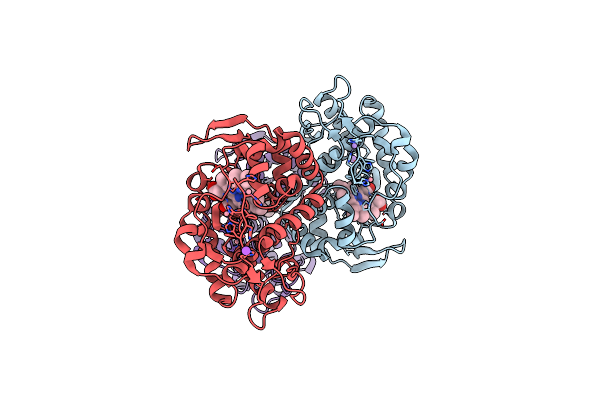
The Crystal Structure Of Engineered Cytochrome C Peroxidase From Saccharomyces Cerevisiae With Trp51 To S-Trp51 And Trp191Phe Modifications
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-06-16 Classification: OXIDOREDUCTASE Ligands: HEM, EDO |
 |
The Crystal Structure Of Engineered Cytochrome C Peroxidase From Saccharomyces Cerevisiae With A His175Me-His Proximal Ligand Substitution
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-02-12 Classification: OXIDOREDUCTASE Ligands: HEM, CO, MN, NA |
 |
Crystal Structure Of Bh32 Alkylated With The Mechanistic Inhibitor 2-Bromoacetophenone
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2019-06-05 Classification: HYDROLASE Ligands: AC0 |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-06-05 Classification: HYDROLASE Ligands: CA |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2019-06-05 Classification: HYDROLASE Ligands: PGE, AC0, SO4, EDO, MG |
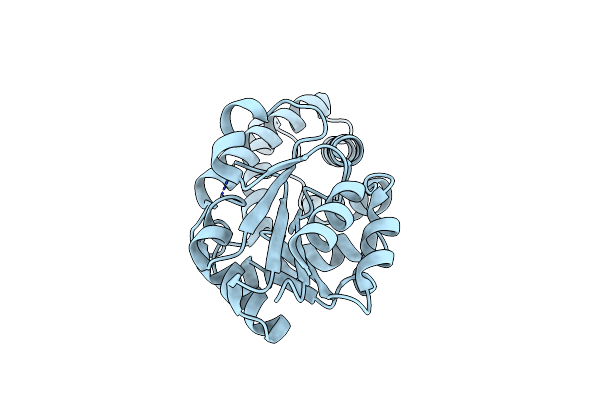 |
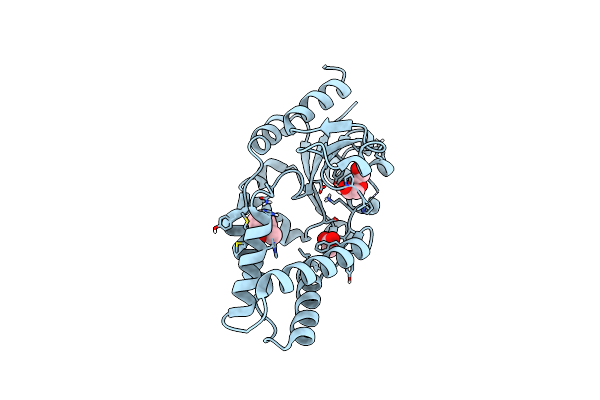
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-06-05 Classification: HYDROLASE |
 |
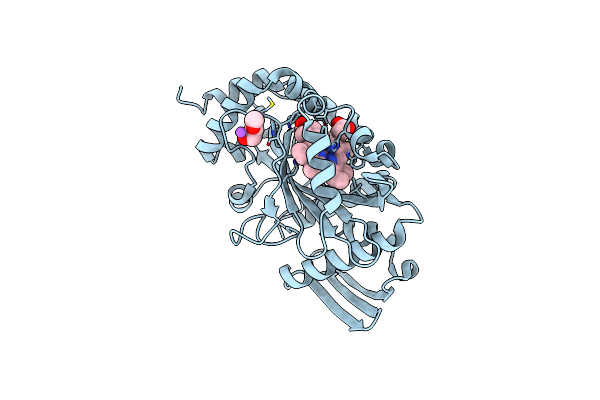
Crystal Structure Of Oe1.3 Alkylated With The Mechanistic Inhibitor 2-Bromoacetophenone
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-06-05 Classification: HYDROLASE Ligands: AC0, ACT, PEG |
 |
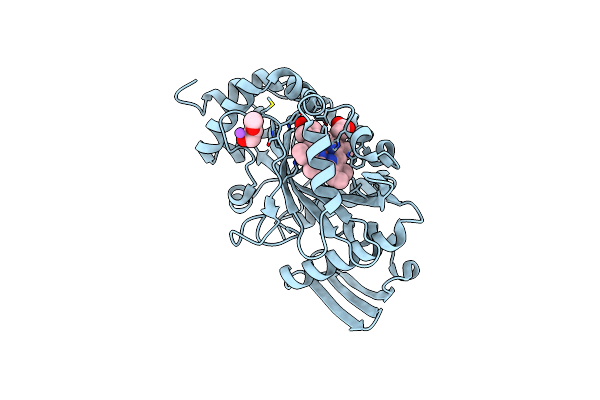
Crystal Structure Of The Alpha Subunit Of Heme Dependent Oxidative N-Demethylase (Hodm)
Organism: Pseudomonas mendocina (strain ymp)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-11-09 Classification: OXIDOREDUCTASE Ligands: HEM, ETX, NA |
 |
Crystal Structure Of The Alpha Subunit Of Heme Dependent Oxidative N-Demethylase (Hodm) In Complex With The Dimethylamine Substrate
Organism: Pseudomonas mendocina
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2016-11-09 Classification: OXIDOREDUCTASE Ligands: HEM, DMN, PEG, NA |
 |
Crystal Structure Of The Alpha Subunit Of Heme Dependent Oxidative N-Demethylase (Hodm) In Complex With The Dimethylamine Substrate
Organism: Pseudomonas mendocina
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-11-09 Classification: OXIDOREDUCTASE Ligands: HEM, DMN, NO, PEG, NA |
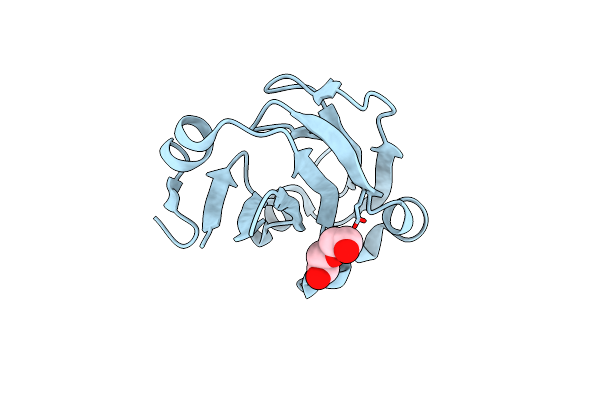 |
Crystal Structure Of Glycine Cleavage Protein H-Like (Gcvh-L) From Streptococcus Pyogenes
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-07-29 Classification: TRANSPORT PROTEIN Ligands: 1PE |
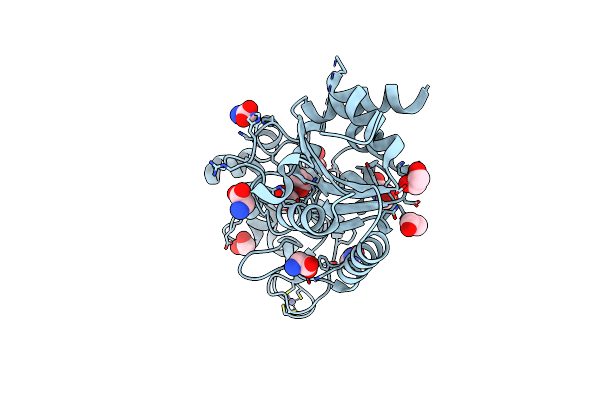 |
Crystal Structure Of The Adp-Ribosylating Sirtuin (Sirtm) From Streptococcus Pyogenes (Apo Form)
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2015-07-29 Classification: TRANSFERASE Ligands: GLY, ZN, EDO |
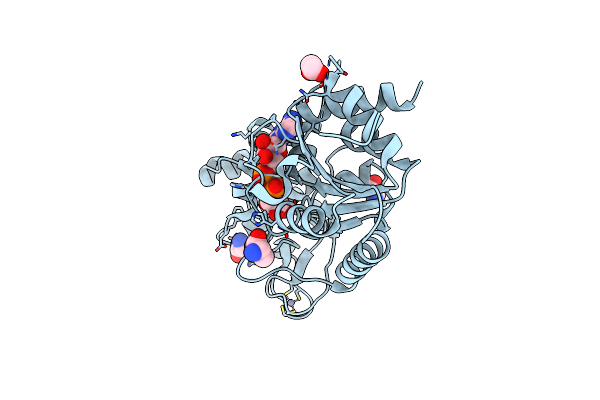 |
Crystal Structure Of The Adp-Ribosylating Sirtuin (Sirtm) From Streptococcus Pyogenes In Complex With Adp-Ribose
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-07-29 Classification: TRANSFERASE Ligands: GLY, ALA, ZN, APR, EDO |
 |
Crystal Structure Of The Adp-Ribosylating Sirtuin (Sirtm) From Streptococcus Pyogenes In Complex With Nad
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2015-07-29 Classification: TRANSFERASE Ligands: GLY, ZN, NAD, EDO |

