Search Count: 54
 |
Polyethylene 600-Bound Form Of P450 Cyp125A3 Mutant From Myobacterium Smegmatis - W83Y
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2015-11-18 Classification: OXIDOREDUCTASE Ligands: HEM, PO4, CIT, 1PE |
 |
X-Ray Crystal Structure Of Cyp119 From Sulfolobus Acidocaldarius, Complexed With 4-(4-Chlorophenyl)Imidazole
Organism: Sulfolobus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-02-18 Classification: OXIDOREDUCTASE Ligands: HEM, CPZ |
 |
X-Ray Crystal Structure Of Cyp142A2 From Mycobacterium Smegmatis, Complexed With Cholesterol Sulfate.
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-09-17 Classification: OXIDOREDUCTASE Ligands: HEM, C3S, GOL |
 |
X-Ray Crystal Structure Of Ligand Free Cyp142A2 From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2014-09-17 Classification: OXIDOREDUCTASE Ligands: HEM |
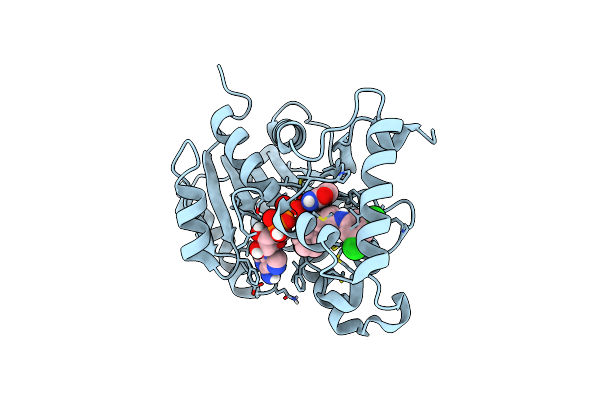 |
Crystal Structure Of Mycobacterium Tuberculosis Enoyl Reductase (Inha) Complexed With 1-Cyclohexyl-N-(3,5-Dichlorophenyl)-5-Oxopyrrolidine-3-Carboxamide
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2014-08-20 Classification: OXIDOREDUCTASE Ligands: NAD, 641 |
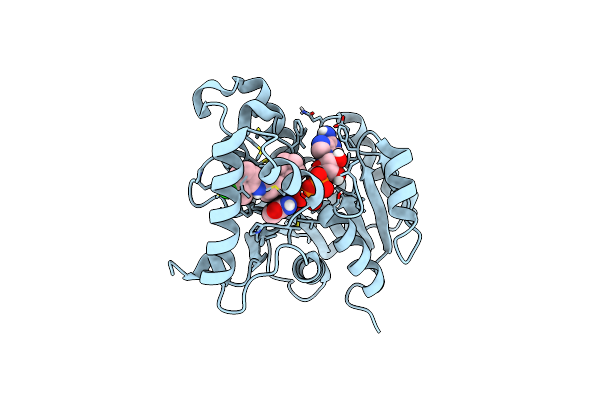 |
Crystal Structure Of Mycobacterium Tuberculosis Enoyl Reductase (Inha) Complexed With N-(3-Chloro-2-Methylphenyl)-1-Cyclohexyl- 5-Oxopyrrolidine-3-Carboxamide
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2014-08-20 Classification: OXIDOREDUCTASE Ligands: NAD, 468 |
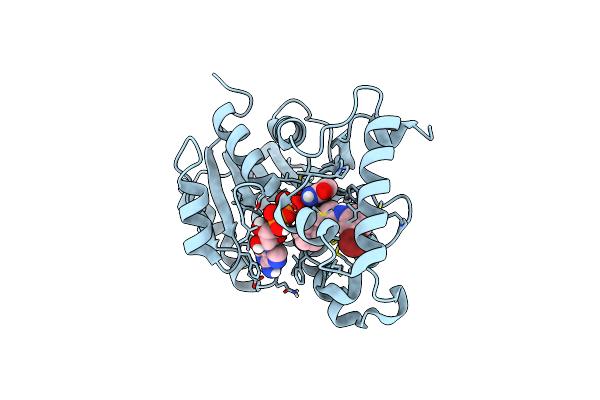 |
Crystal Structure Of Mycobacterium Tuberculosis Enoyl Reductase (Inha) Complexed With N-(3-Bromophenyl)-1-Cyclohexyl-5-Oxopyrrolidine-3-Carboxamide, Refined With New Ligand Restraints
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2014-08-13 Classification: OXIDOREDUCTASE Ligands: NAD, 665 |
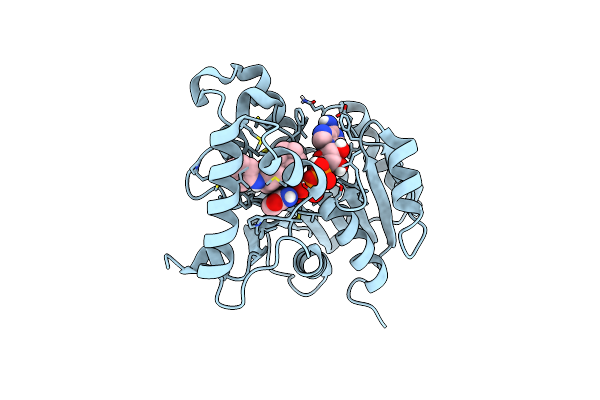 |
Crystal Structure Of Mycobacterium Tuberculosis Enoyl Reductase (Inha) Complexed With 1-Cyclohexyl-5-Oxo-N-Phenylpyrrolidine-3-Carboxamide, Refined With New Ligand Restraints
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2014-07-30 Classification: OXIDOREDUCTASE Ligands: NAD, 566 |
 |
Crystal Structure Of Mycobacterium Tuberculosis Enoyl Reductase Complexed With N-(5-Chloro-2-Methylphenyl)-1-Cyclohexyl-5-Oxopyrrolidine-3-Carboxamide
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-07-30 Classification: OXIDOREDUCTASE Ligands: NAD, 744 |
 |
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2013-02-27 Classification: OXIDOREDUCTASE Ligands: HEM, K2B, MG |
 |
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Release Date: 2013-02-27 Classification: OXIDOREDUCTASE Ligands: HEM, SO4, EDO |
 |
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-02-27 Classification: OXIDOREDUCTASE Ligands: EDO, HEM, CL |
 |
X-Ray Structure Of The Substrate-Free Mycobacterium Tuberculosis Cytochrome P450 Cyp125, Alternative Crystal Form
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2010-11-10 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
X-Ray Structure Of Mycobacterium Tuberculosis Cyp125 Bound To The Reverse Type I Inhibitor
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-11-10 Classification: OXIDOREDUCTASE Ligands: HEM, RT8 |
 |
X-Ray Structure Of The Substrate-Free Mycobacterium Tuberculosis Cytochrome P450 Cyp125
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2010-11-10 Classification: OXIDOREDUCTASE Ligands: SO4, HEM |
 |
X-Ray Structure Of Mycobacterium Tuberculosis Cytochrome P450 Cyp125 In Complex With Substrate Cholest-4-En-3-One
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2010-06-16 Classification: OXIDOREDUCTASE Ligands: SO4, K2B, EDO, HEM |
 |
X-Ray Structure Of Mycobacterium Tuberculosis Cytochrome P450 Cyp124 In Complex With Phytanic Acid
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2009-10-06 Classification: OXIDOREDUCTASE Ligands: HEM, VGJ, CA |
 |
X-Ray Structure Of The Substrate-Free Mycobacterium Tuberculosis Cytochrome P450 Cyp124
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-10-06 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
X-Ray Structure Of The Heme-Bound Gaf Domain Of Sensory Histidine Kinase Dost Of Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-03-17 Classification: TRANSFERASE Ligands: HEM, ACT, OXY |
 |
Crystal Structure Of A Cytochrome P450 From The Thermoacidophilic Archaeon Picrophilus Torridus
Organism: Picrophilus torridus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-01-29 Classification: OXIDOREDUCTASE Ligands: SO4, HEM |

