Search Count: 15
 |
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2021-12-29 Classification: OXIDOREDUCTASE Ligands: PGE, PG4 |
 |
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2021-12-29 Classification: OXIDOREDUCTASE Ligands: PG4 |
 |
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2021-12-29 Classification: OXIDOREDUCTASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2019-08-07 Classification: HYDROLASE |
 |
Extremely Stable Monomeric Variant Of Human Cystatin C With Single Amino Acid Substitution
Organism: Homo sapiens
Method: SOLUTION NMR, SOLUTION SCATTERING Release Date: 2019-07-31 Classification: HYDROLASE |
 |
Organism: Trametes sanguinea
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2017-11-01 Classification: OXIDOREDUCTASE Ligands: CU, PER |
 |
Organism: Pycnoporus coccineus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-11-01 Classification: OXIDOREDUCTASE Ligands: CU, PER, ME2, IPH, NA, MLI |
 |
Crystal Structure Of Laccases From Pycnoporus Sanguineus, Izoform Ii, Monoclinic
Organism: Trametes sanguinea
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2017-11-01 Classification: OXIDOREDUCTASE Ligands: CU, PER, PEG, FMT, NAG |
 |
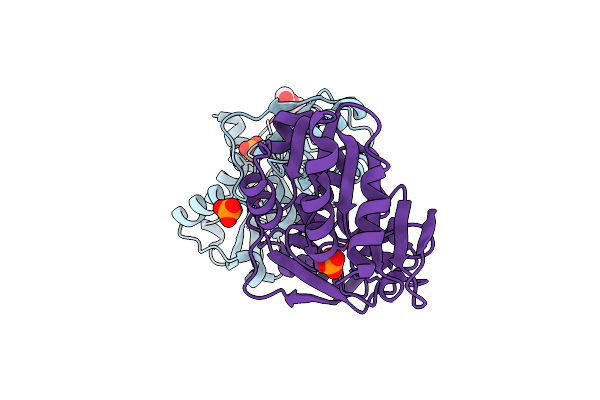
Organism: Francisella tularensis
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2013-10-02 Classification: ISOMERASE Ligands: 5RP, ARF, CL |
 |
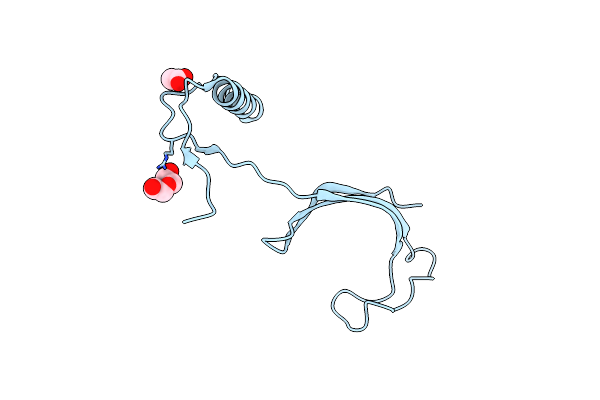
Organism: Francisella tularensis subsp. tularensis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-02-13 Classification: ISOMERASE Ligands: CL, BR, PO4, PG4 |
 |
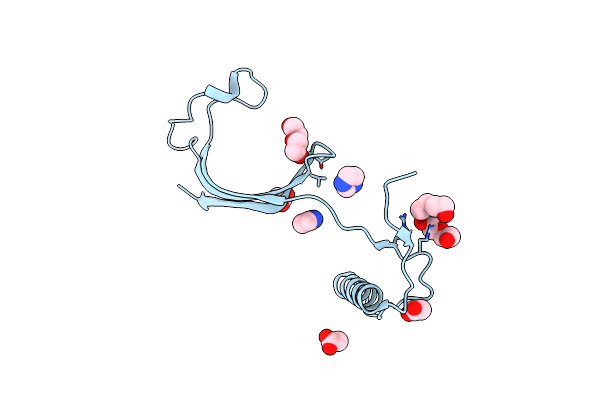
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2012-08-01 Classification: hydrolase inhibitor Ligands: PEG, ACT |
 |
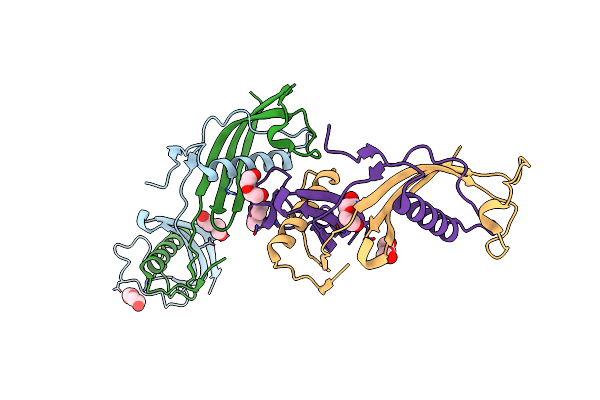
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2012-05-30 Classification: HYDROLASE INHIBITOR Ligands: PEG, IMD, ACT, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2012-02-29 Classification: HYDROLASE INHIBITOR Ligands: PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2011-12-21 Classification: HYDROLASE INHIBITOR Ligands: PEG, ACT |
 |
Hinge-Loop Mutation Can Be Used To Control 3D Domain Swapping And Amyloidogenesis Of Human Cystatin C
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2010-12-01 Classification: HYDROLASE INHIBITOR Ligands: SO4 |

