Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Blastobotrys adeninivorans
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-02-17 Classification: LYASE Ligands: 4NC, K, CO |
 |
Crystal Structure Of The Pea Pathogenicity Protein 2 From Madurella Mycetomatis
Organism: Madurella mycetomatis
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2021-02-17 Classification: LYASE Ligands: K, CA |
 |
Crystal Structure Of The Pea Pathogenicity Protein 2 From Madurella Mycetomatis Complexed With 4-Nitrocatechol
Organism: Madurella mycetomatis
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2021-02-17 Classification: LYASE Ligands: 4NC, K, CA |
 |
Organism: Blastobotrys adeninivorans
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-02-17 Classification: LYASE Ligands: K |
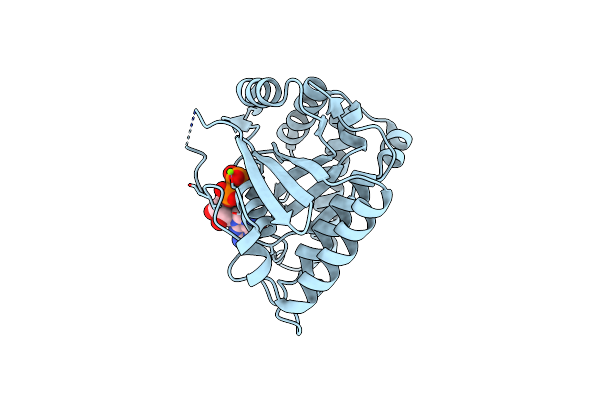 |
Crystal Structure Of Toc33 From Arabidopsis Thaliana, Dimerization Deficient Mutant R130A
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2008-06-24 Classification: HYDROLASE Ligands: MG, GDP |
 |
Crystal Structure Of Toc34 From Pisum Sativum In Complex With Mg2+ And Gmppnp
Organism: Pisum sativum
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2008-04-01 Classification: HYDROLASE Ligands: MG, GNP, PGE, GOL |
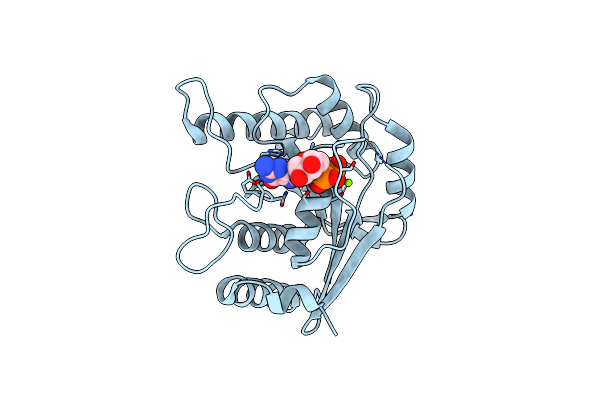 |
Crystal Structure Of Toc33 From Arabidopsis Thaliana In Complex With Gdp And Mg2+
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2008-04-01 Classification: HYDROLASE Ligands: MG, GDP |
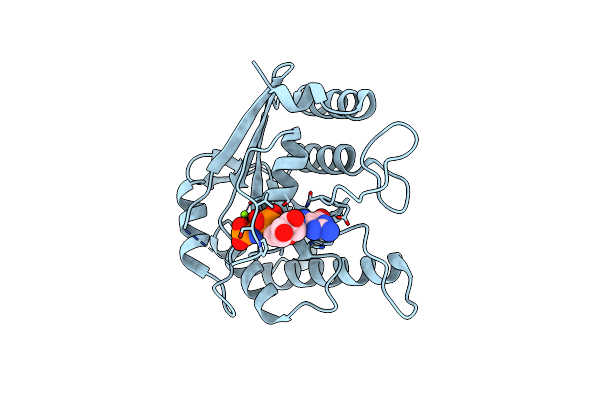 |
Crystal Structure Of Toc33 From Arabidopsis Thaliana In Complex With Mg2+ And Gmppnp
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2008-04-01 Classification: HYDROLASE Ligands: MG, GNP |