Search Count: 17
 |
Structure Of Native Leukocyte Myeloperoxidase In Complex With A Truncated Version Of The Staphylococcal Peroxidase Inhibitor Spin And Selenocyanate At Ph 5.5
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: SEK, HEM, NAG, CA |
 |
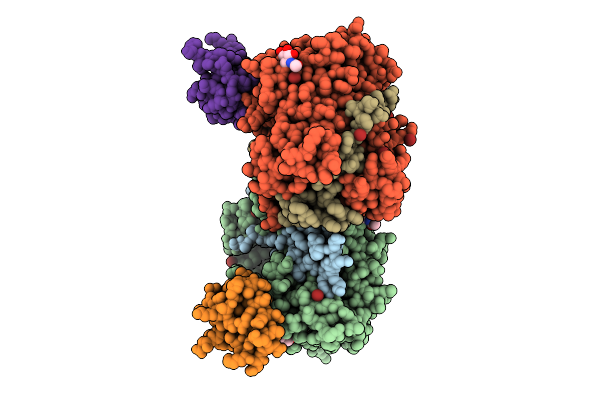
Structure Of Native Leukocyte Myeloperoxidase In Complex With A Truncated Version Of The Staphylococcal Peroxidase Inhibitor Spin And Thiocyanate At Ph 7.5
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: NAG, CA, SCN, HEM |
 |
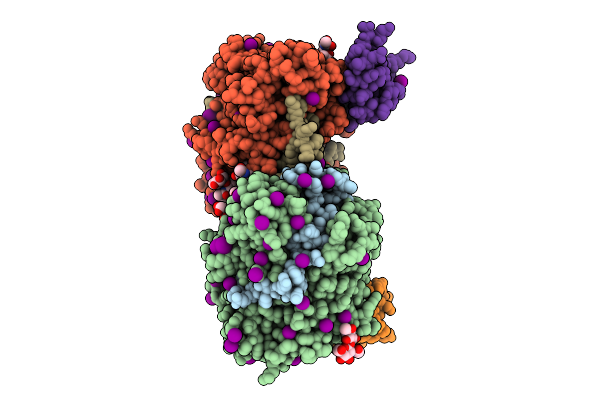
Structure Of Native Leukocyte Myeloperoxidase In Complex With A Truncated Version Of The Staphylococcal Peroxidase Inhibitor Spin And Bromide At Ph 7.5
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: HEM, BR, NAG, CA |
 |
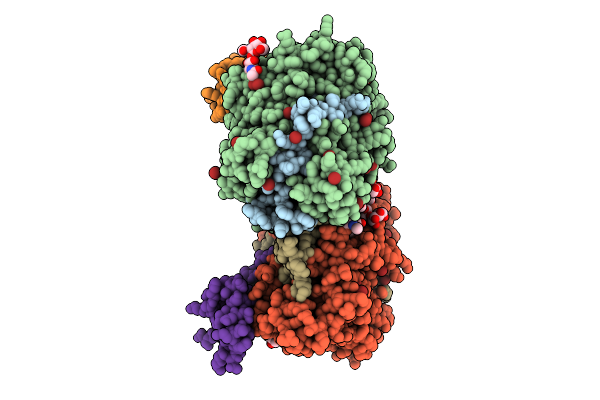
Structure Of Native Leukocyte Myeloperoxidase In Complex With A Truncated Version Of The Staphylococcal Peroxidase Inhibitor Spin And Iodide At Ph 5.5
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: IOD, CA, HEM |
 |
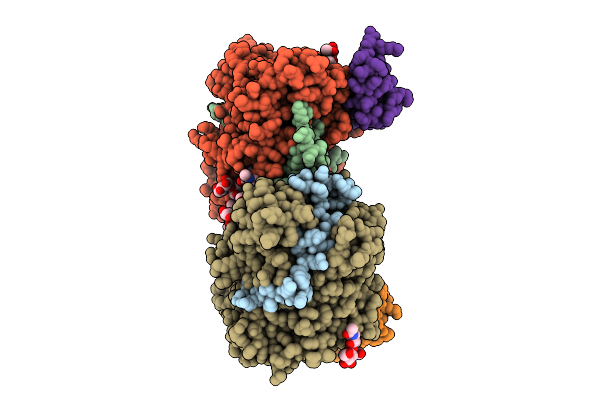
Structure Of Native Leukocyte Myeloperoxidase In Complex With The Staphylococcal Peroxidase Inhibitor Spin And Bromide At Ph 5.5
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: NAG, BR, CA, HEM |
 |
Structure Of Native Leukocyte Myeloperoxidase In Complex With A Truncated Version Of The Staphylococcal Peroxidase Inhibitor Spin And Chloride At Ph 5.5
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: HEM, IOD, CA, CL, NAG |
 |
Organism: Paenibacillus alvei
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-10-02 Classification: ISOMERASE Ligands: PEG |
 |
Structure Of Coproheme Decarboxylase From Corynebacterium Diphteriae In Complex With Monovinyl Monopropionyl Deuteroheme
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-04-22 Classification: OXIDOREDUCTASE Ligands: VOV |
 |
Structure Of Coproheme Decarboxylase From Corynebacterium Diphteriae In Complex With Coproheme
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2020-04-22 Classification: OXIDOREDUCTASE Ligands: FEC |
 |
Structure Of Coproheme Decarboxylase From Listeria Monocytogenes In Complex With Iron Coproporphyrin Iii
Organism: Listeria monocytogenes egd-e
Method: X-RAY DIFFRACTION Release Date: 2019-07-10 Classification: OXIDOREDUCTASE |
 |
Structure Of Coproheme Decarboxylase From Listeria Monocytogenes During Turnover
Organism: Listeria monocytogenes serovar 1/2a (strain atcc baa-679 / egd-e)
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2019-07-10 Classification: OXIDOREDUCTASE |
 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2018-09-12 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2017-04-05 Classification: OXIDOREDUCTASE Ligands: NAG, HEM, CA, CL, PEG, TRS |
 |
Organism: Magnaporthe oryzae (strain 70-15 / atcc mya-4617 / fgsc 8958)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-06-22 Classification: OXIDOREDUCTASE Ligands: HEM, FLC |
 |
Organism: Magnaporthe oryzae (strain 70-15 / atcc mya-4617 / fgsc 8958)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-06-22 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Magnaporthe oryzae (strain 70-15 / atcc mya-4617 / fgsc 8958)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-06-22 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Magnaporthe oryzae (strain 70-15 / atcc mya-4617 / fgsc 8958)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-09-02 Classification: OXIDOREDUCTASE Ligands: 522, OH |

