Search Count: 24
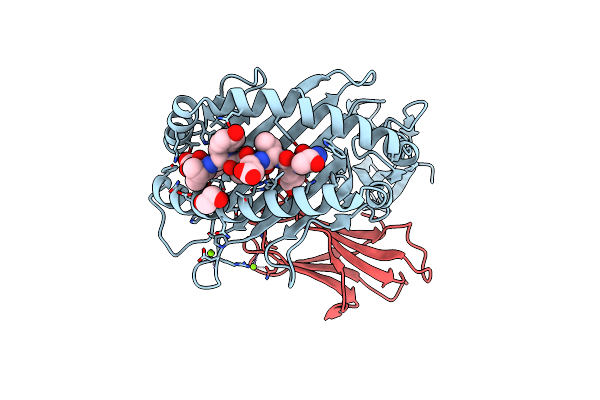 |
Crystal Structure Of Hla B*18:01 In Complex With Tevetyvl, An 8-Mer Epitope From Influenza A
Organism: Homo sapiens, Influenza a virus (a/memphis/18/1978(h3n2))
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2024-05-08 Classification: IMMUNE SYSTEM Ligands: EDO, MG |
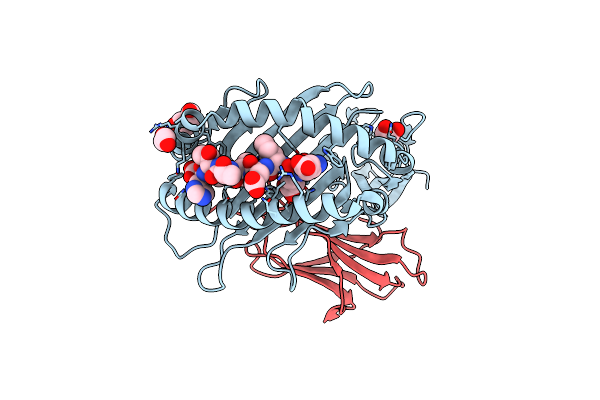 |
Crystal Structure Of Hla B*18:01 In Complex With Eeieitthf, An 9-Mer Epitope From Influenza A
Organism: Homo sapiens, Influenza a virus (a/memphis/18/1978(h3n2))
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-05-08 Classification: IMMUNE SYSTEM Ligands: PEG, EDO |
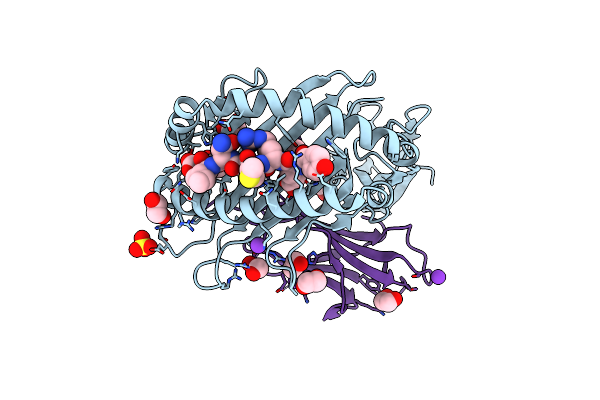 |
Crystal Structure Of Hla B*18:01 In Complex With Yermcnil, An 8-Mer Epitope From Influenza A
Organism: Homo sapiens, Influenza a virus (a/x-31(h3n2))
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-05-08 Classification: IMMUNE SYSTEM Ligands: EDO, MG, K, SO4, NO3 |
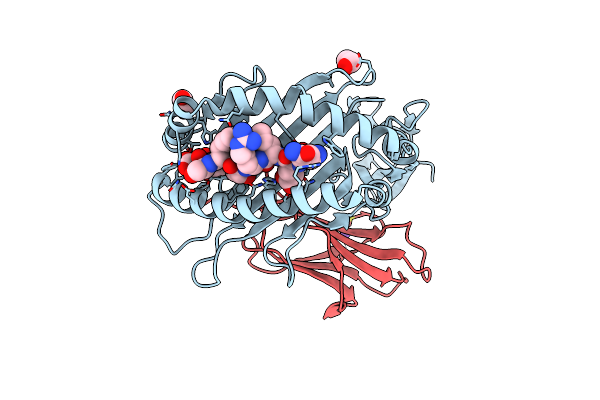 |
Crystal Structure Of Hla B*18:01 In Complex With Qeirtfsf, An 8-Mer Epitope From Influenza A
Organism: Homo sapiens, Influenza a virus (a/x-31(h3n2))
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2024-05-08 Classification: IMMUNE SYSTEM Ligands: EDO, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-12-06 Classification: CYTOKINE Ligands: UTF |
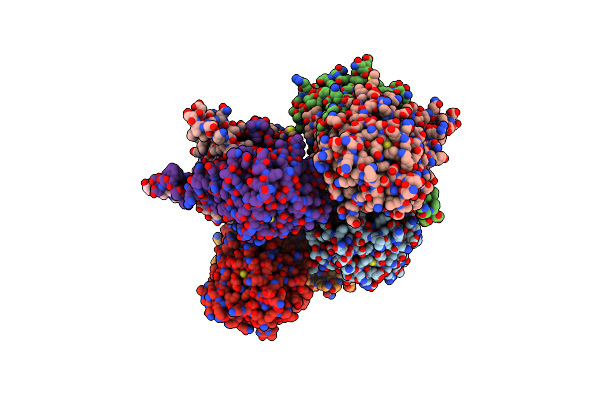 |
Structure Of Lactobacillus Salivarius (Ls) Bile Salt Hydrolase(Bsh) In Complex With Glycocholate (Gca)
Organism: Ligilactobacillus salivarius
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-03-08 Classification: HYDROLASE Ligands: GCH |
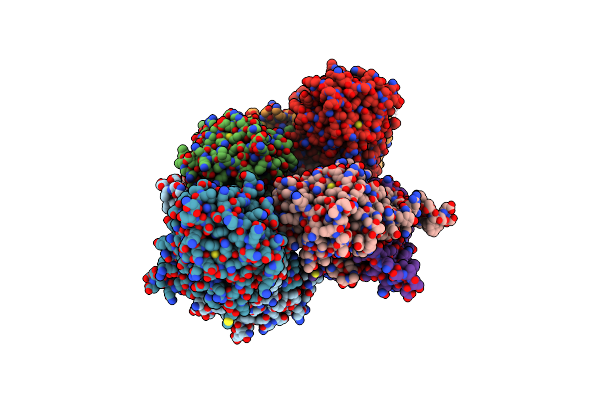 |
Structure Of Lactobacillus Salivarius (Ls) Bile Salt Hydrolase(Bsh) In Complex With Taurocholate (Tca)
Organism: Ligilactobacillus salivarius
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-03-08 Classification: HYDROLASE Ligands: TCH |
 |
Organism: Mus musculus, Human alphaherpesvirus 1 strain kos
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: VIRAL PROTEIN |
 |
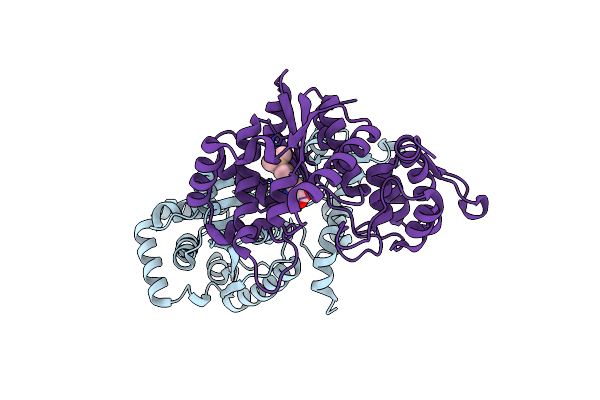
Organism: Human alphaherpesvirus 1 strain kos, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: VIRAL PROTEIN/Immune System |
 |
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2021-03-31 Classification: LIGASE Ligands: TSQ |
 |
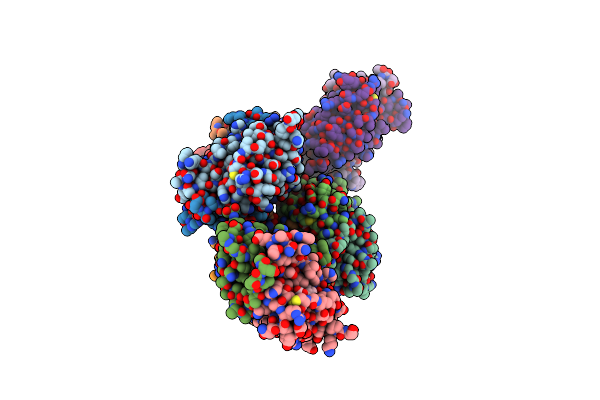
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2021-03-31 Classification: LIGASE |
 |
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-12-23 Classification: Biotin-binding protein Ligands: BTN |
 |
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-12-23 Classification: Biotin-binding protein Ligands: BTN |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-09-09 Classification: FLUORESCENT PROTEIN Ligands: GOL, CL, NA |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-06-13 Classification: SIGNALING PROTEIN |
 |
Organism: Caenorhabditis elegans
Method: SOLUTION NMR Release Date: 2013-10-23 Classification: UNKNOWN FUNCTION |
 |
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-05-01 Classification: UNKNOWN FUNCTION |
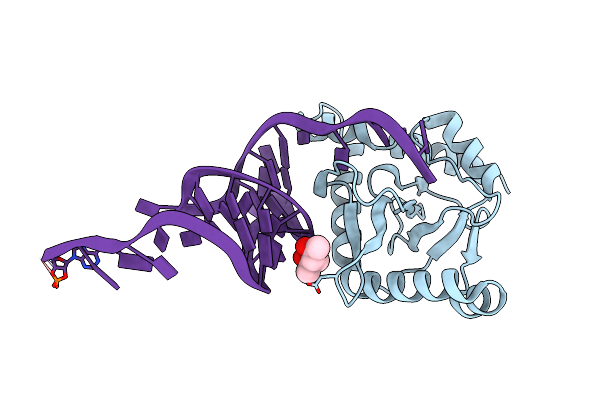 |
New Insights Into The Mechanism Of Bacterial Type Iii Toxin-Antitoxin Systems: Selective Toxin Inhibition By A Non-Coding Rna Pseudoknot
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-12-26 Classification: TOXIN/ANTITOXIN Ligands: MPD |
 |
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-06-13 Classification: biotin-binding protein Ligands: BTN, CL |
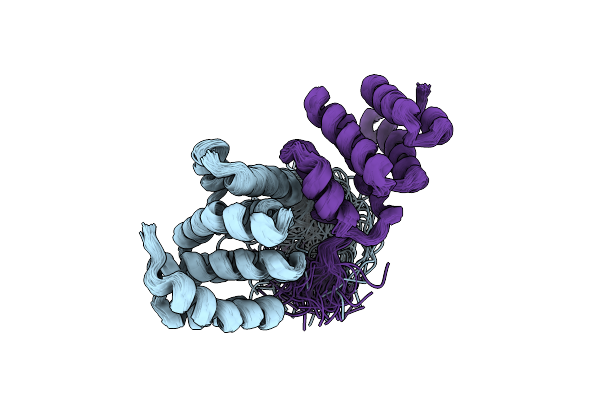 |
Organism: Homo sapiens, Mus musculus
Method: SOLUTION NMR Release Date: 2011-03-16 Classification: APOPTOSIS |

