Search Count: 1,404
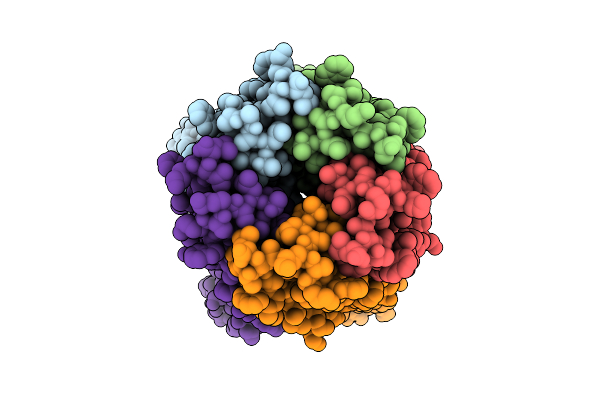 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: DE NOVO PROTEIN Ligands: IOD |
 |
Organism: Chroococcidiopsis thermalis pcc 7203
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: PHOTOSYNTHESIS Ligands: CL0, F6C, CLA, PQN, SF4, BCR, LHG, LMT, LMG, LFA, CA |
 |
Cryo-Em Structure Of Slc30A10 In Mn2+-Bound State, Determined In Inward-Facing Conformation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN Ligands: MN |
 |
Cryo-Em Structure Of Slc30A10, Determined In Asymmetric Conformations-One Subunit In An Inward-Facing Mn2+-Bound And The Other In An Outward-Facing Mn2+-Unbound Conformation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN Ligands: MN |
 |
Cryo-Em Structure Of Slc30A10 In The Absence Of Mn2+, Determined In Inward-Facing Conformation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of Wild-Type Egfr In Complex With The Reversible Inhibitor Sevabertinib (Bay 2927088)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: SIN, CL, DMS, EDO, A1JBI |
 |
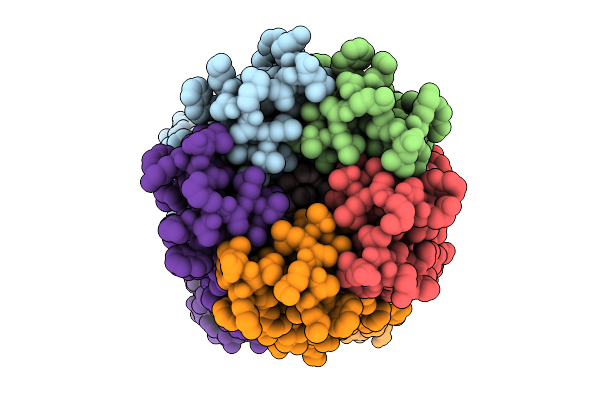
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE Ligands: A1IQT, ZN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: DE NOVO PROTEIN Ligands: LMT |
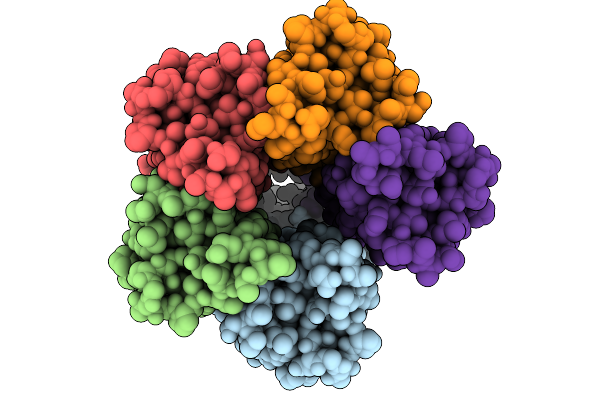 |
Cryo-Em Structure Of A De Novo Designed Voltage-Gated Anion Channel (Dvgac)
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: DE NOVO PROTEIN |
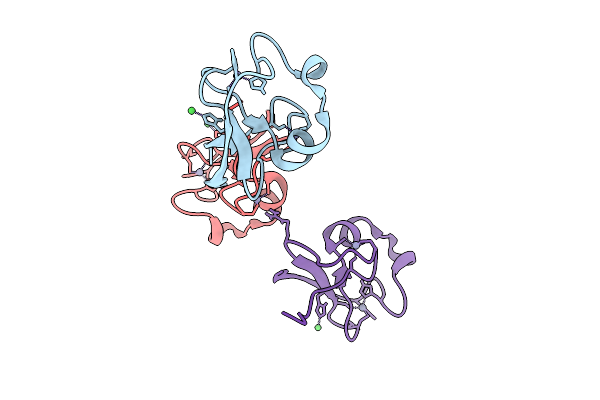 |
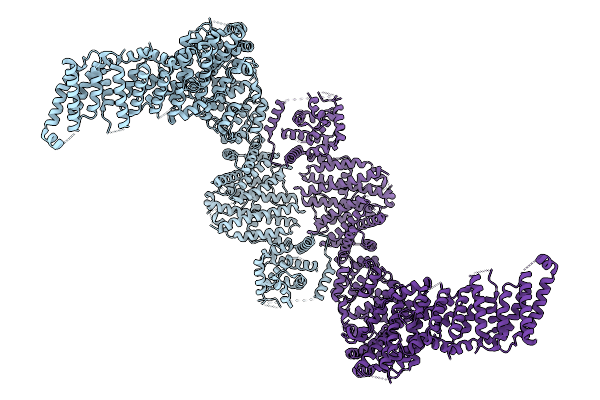
R-Degron Fused Zz-Domain Of The Arabidopsis Thaliana E3 Ubiquitin-Protein Ligase Big
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: LIGASE Ligands: ZN, NI |
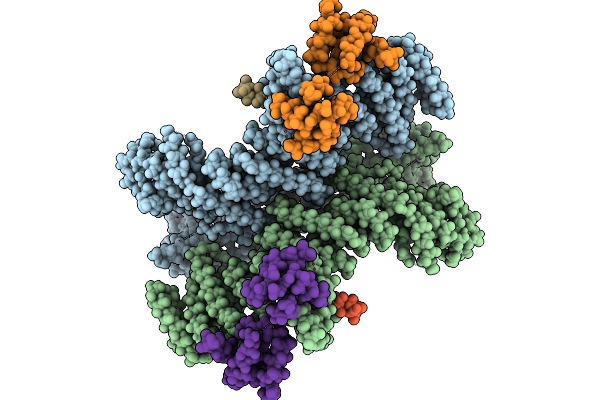 |
Cryo-Em Structure Of The Core Of The Arabidopsis Thaliana Ubr4/Di19/Calm1 Complex
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE |
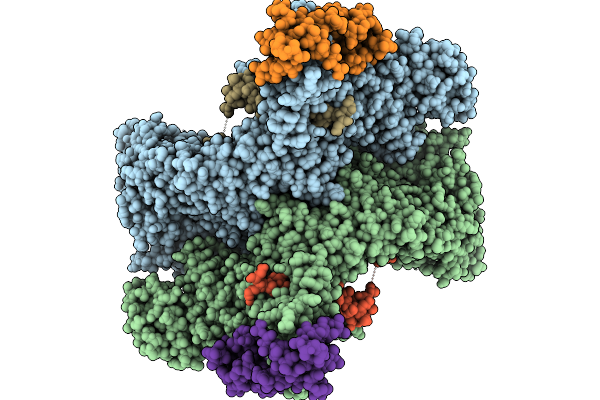 |
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (C-Term Dimer Interface Focused Refinement)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE Ligands: ZN |
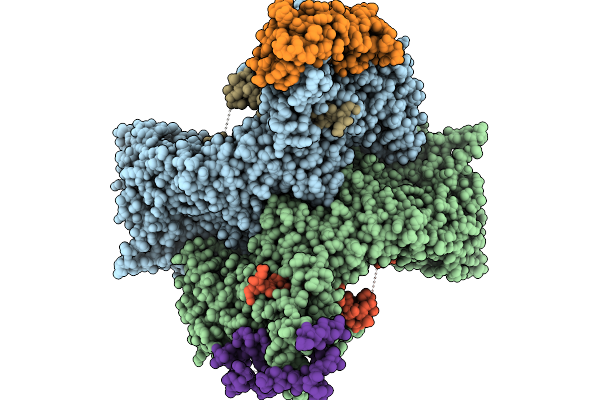 |
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (Calm1 Focused Refinement)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE Ligands: ZN |
 |
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (N-Term Focused Refinement)
|
 |
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (Bp Focused Refinement)
|
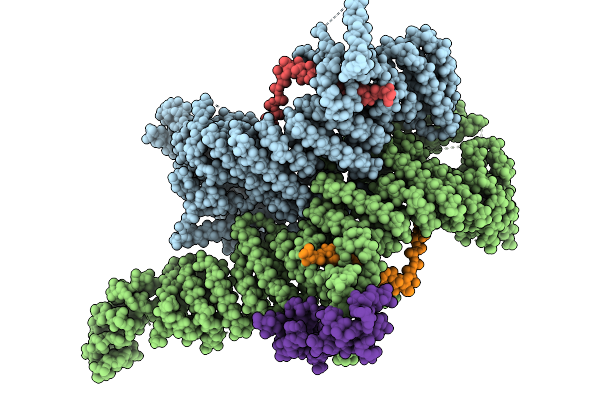 |
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (C-Term Focused Refinement)
|
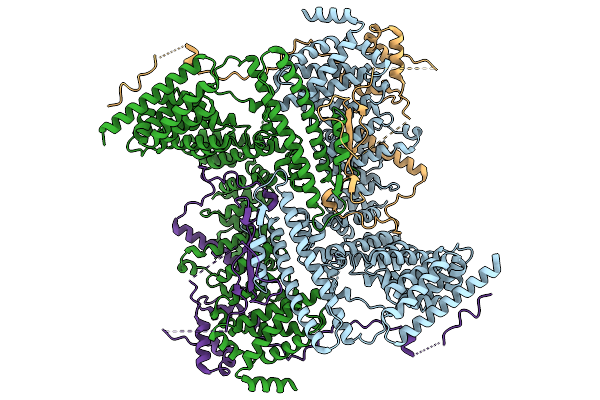 |
Cryo-Em Structure Of The C. Elegans Ubr4/Kcmf1 Complex (C-Term Dimer Interface Focused Refinement)
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE Ligands: ZN |
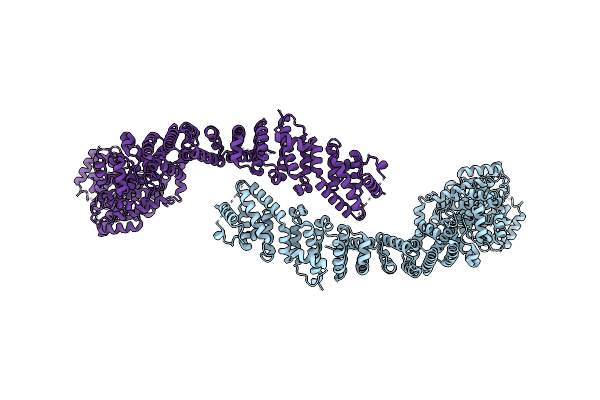 |
Cryo-Em Structure Of The C. Elegans Ubr4/Kcmf1 Complex (N-Term Focused Refinement)
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE |
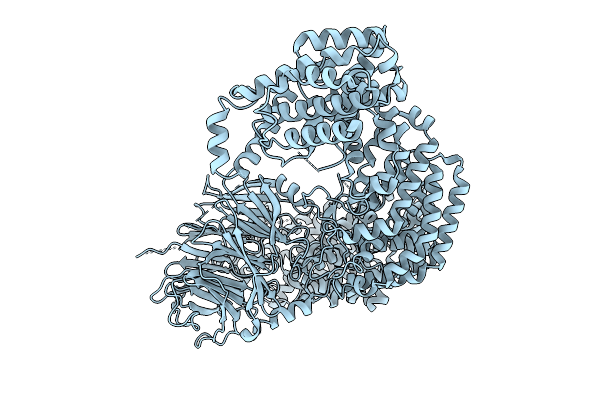 |
Cryo-Em Structure Of The C. Elegans Ubr4/Kcmf1 Complex (Side Focused Refinement)
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN |

