Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PROTEIN FIBRIL |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PROTEIN FIBRIL |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PROTEIN FIBRIL |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PROTEIN FIBRIL |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PROTEIN FIBRIL |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PROTEIN FIBRIL |
 |
Crystal Structure Of N-Terminal Domain Of N-Methyl-D-Aspartate Receptor Subunit Nr1 In Complex With Patient-Derived Antibody
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN |
 |
Structure Of Human Pyruvate Dehydrogenase Kinase 2 Complexed With Compound 6
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-06-19 Classification: TRANSFERASE Ligands: A1L16 |
 |
Structure Of Human Pyruvate Dehydrogenase Kinase 2 Complexed With Compound 16
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2024-06-19 Classification: TRANSFERASE Ligands: A1L17 |
 |
Optimization Efforts For Identification Of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction Ihhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2024-05-22 Classification: PEPTIDE BINDING PROTEIN Ligands: A1LVB, ACT, SO4 |
 |
Optimization Efforts For Identification Of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction (Ppi) Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2024-05-22 Classification: PEPTIDE BINDING PROTEIN Ligands: A1LVC, ACT, SO4 |
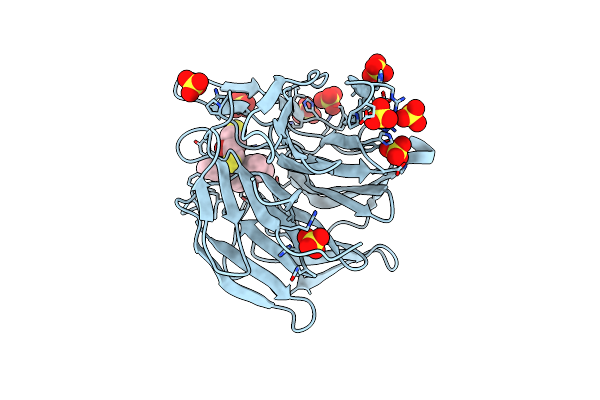 |
Methyl And Fluorine Effects In Novel Orally Bioavailable Keap1/Nrf2 Ppi Inhibitor For Treatment Of Chronic Kidney Disease
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-06-14 Classification: PEPTIDE BINDING PROTEIN Ligands: SGO, DTT, SO4 |
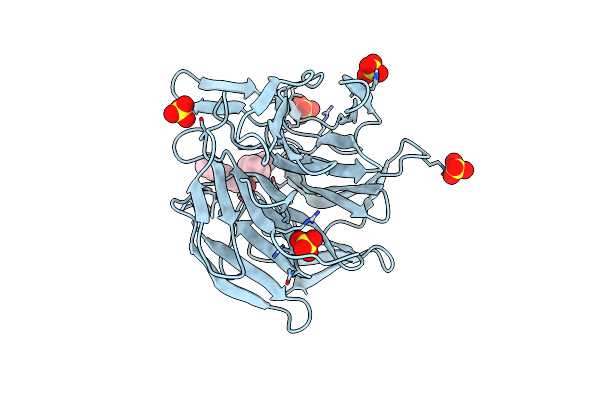 |
Methyl And Fluorine Effects In Novel Orally Bioavailable Keap1/Nrf2 Ppi Inhibitor For Treatment Of Chronic Kidney Disease
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-06-14 Classification: PEPTIDE BINDING PROTEIN Ligands: SIU, SO4 |
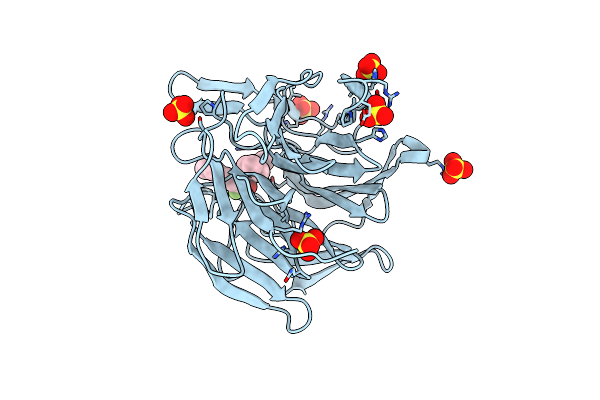 |
Methyl And Fluorine Effects In Novel Orally Bioavailable Keap1/Nrf2 Ppi Inhibitor For Treatment Of Chronic Kidney Disease
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2023-06-14 Classification: PEPTIDE BINDING PROTEIN Ligands: T6I, SO4 |
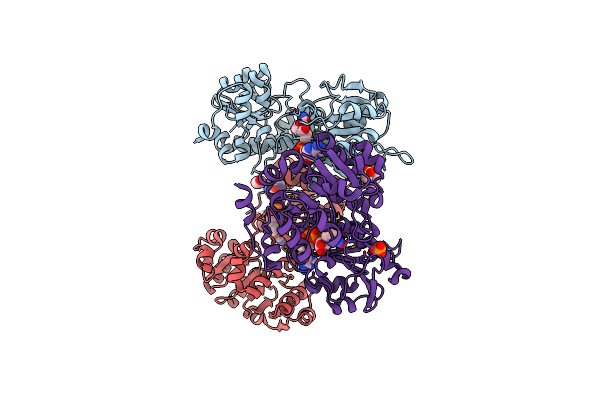 |
Organism: Geobacillus kaustophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-04-05 Classification: OXIDOREDUCTASE Ligands: NAD, EDO, EPE, PO4 |
 |
Organism: Geobacillus kaustophilus
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-04-05 Classification: OXIDOREDUCTASE Ligands: NAD, EDO, PYR |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-06-22 Classification: HYDROLASE |