Search Count: 25
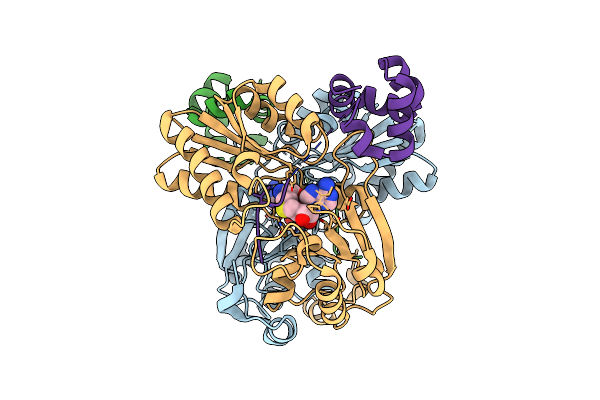 |
Structure Of The Alpha-N-Methyltransferase (Sonm) And Ripp Precursor (Sona With Qsy Deletion) Heteromeric Complex (Bound To Sah)
Organism: Shewanella oneidensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-22 Classification: TRANSFERASE Ligands: SAH, ZN |
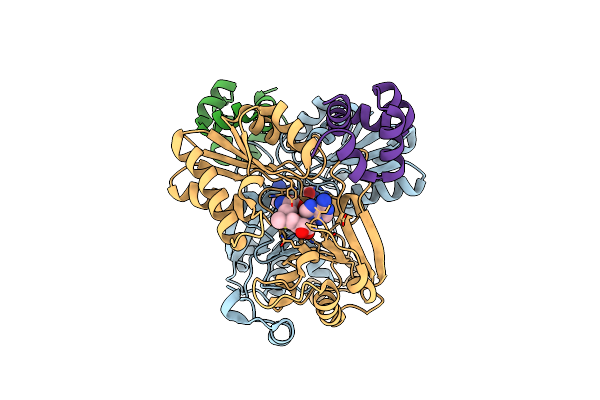 |
Structure Of The Alpha-N-Methyltransferase (Sonm) And Ripp Precursor (Sona With Qsy Deletion) Heteromeric Complex (Bound To Sam)
Organism: Shewanella oneidensis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-11-22 Classification: TRANSFERASE Ligands: SAM, ZN |
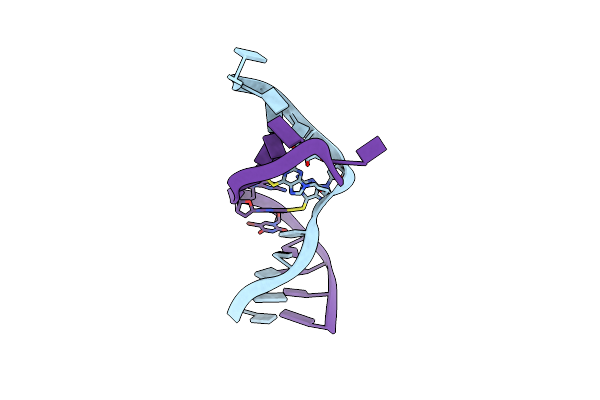 |
Crystal Structure Of A Dna Duplex Cross-Linked By 6-Thioguanine-6-Thioguanine Disulfides
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2019-10-30 Classification: DNA |
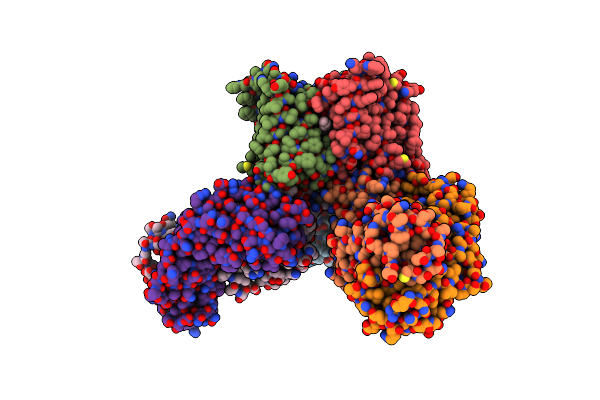 |
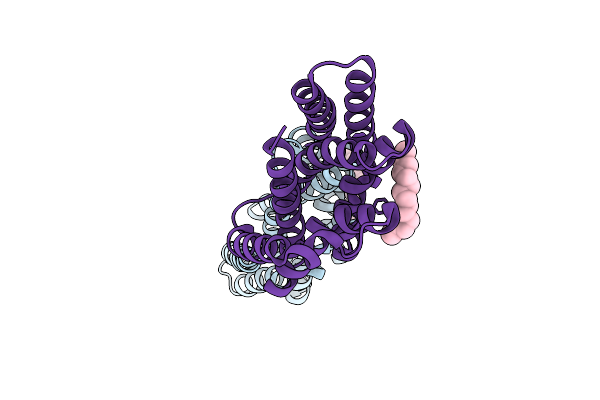
Crystal Structures Of The Tric Trimeric Intracellular Cation Channel Orthologue From Sulfolobus Solfataricus
Organism: Mus musculus, Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2017-01-11 Classification: IMMUNE SYSTEM/MEMBRANE PROTEIN Ligands: GOL, NA, PX4 |
 |
Crystal Structures Of The Tric Trimeric Intracellular Cation Channel Orthologue From Rhodobacter Sphaeroides
Organism: Rhodobacter sphaeroides (strain atcc 17023 / 2.4.1 / ncib 8253 / dsm 158)
Method: X-RAY DIFFRACTION Resolution:3.41 Å Release Date: 2017-01-11 Classification: MEMBRANE PROTEIN Ligands: PX4 |
 |
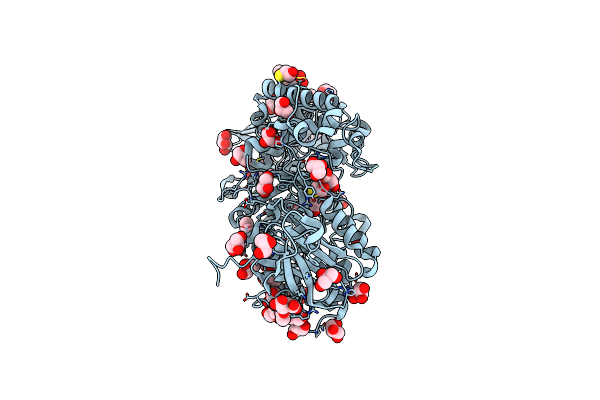
Structure Of The Starch Branching Enzyme I (Bei) Complexed With Maltopentaose From Oryza Sativa L
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2013-05-08 Classification: TRANSFERASE Ligands: BGC, GOL |
 |
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-09-28 Classification: TRANSFERASE Ligands: PO4, GOL |
 |
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-09-28 Classification: TRANSFERASE Ligands: BME, GOL, EPE, ACT, SIN |
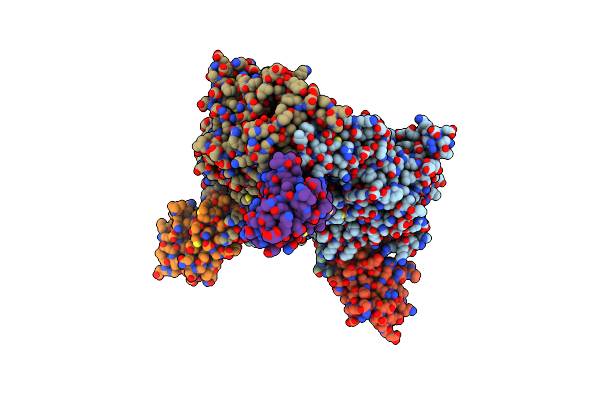 |
Crystal Structure Of The Electron Transfer Complex Between Oxygenase And Ferredoxin In Carbazole 1,9A-Dioxygenase
Organism: Janthinobacterium, Pseudomonas resinovorans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-12-05 Classification: OXIDOREDUCTASE Ligands: FE2, FES |
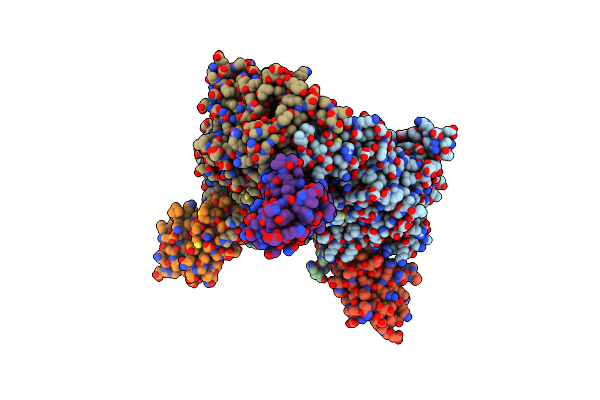 |
The Reduced Complex Between Oxygenase And Ferredoxin In Carbazole 1,9A-Dioxygenase
Organism: Janthinobacterium, Pseudomonas resinovorans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-12-05 Classification: OXIDOREDUCTASE Ligands: FE2, FES |
 |
The Substrate-Bound Complex Between Oxygenase And Ferredoxin In Carbazole 1,9A-Dioxygenase
Organism: Janthinobacterium, Pseudomonas resinovorans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-12-05 Classification: OXIDOREDUCTASE Ligands: FE2, FES, 9CA |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2006-06-06 Classification: HYDROLASE Ligands: CL, PO4 |
 |
Crystal Structure Of The Terminal Oxygenase Component Of Carbazole 1,9A-Dioxygenase, A Non-Heme Iron Oxygenase System Catalyzing The Novel Angular Dioxygenation For Carbazole And Dioxin
Organism: Janthinobacterium sp. j3
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2005-08-23 Classification: OXIDOREDUCTASE Ligands: FE2, FES |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2005-03-29 Classification: OXIDOREDUCTASE Ligands: FE2, FES, OXY |
 |
Crystal Structure Of Ferredoxin Component Of Carbazole 1,9A-Dioxygenase Of Pseudomonas Resinovorans Strain Ca10
Organism: Pseudomonas resinovorans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2005-03-01 Classification: OXIDOREDUCTASE Ligands: FE, FES, H2S |
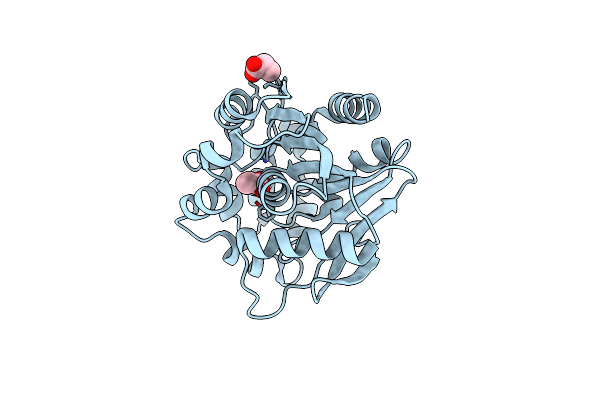 |
Crystal Structure Of A Meta-Cleavage Product Hydrolase (Cumd) Complexed With Propionate
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2004-09-14 Classification: HYDROLASE Ligands: PPI |
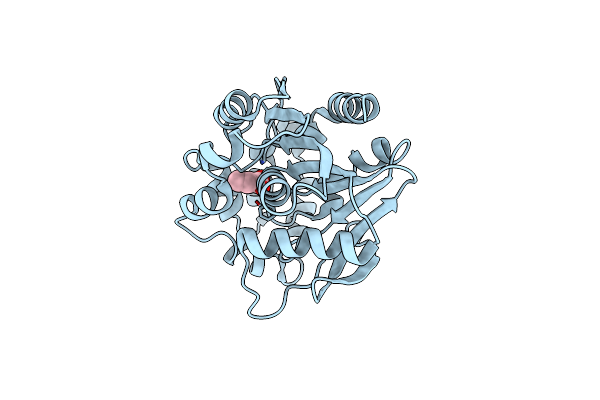 |
Crystal Structure Of A Meta-Cleavage Product Hydrolase (Cumd) Complexed With N-Butyrate
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2004-09-14 Classification: HYDROLASE Ligands: BUA |
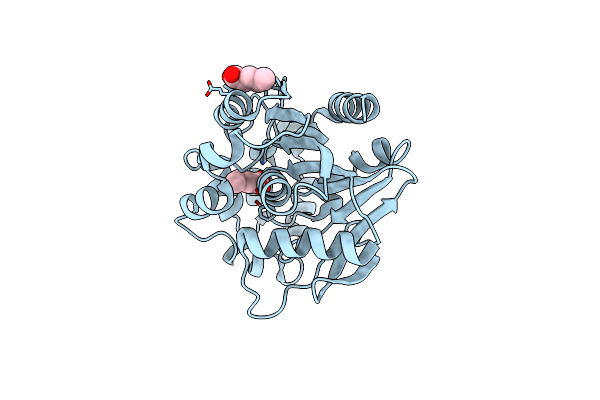 |
Crystal Structure Of A Meta-Cleavage Product Hydrolase (Cumd) Complexed With N-Valerate
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2004-09-14 Classification: HYDROLASE Ligands: LEA |
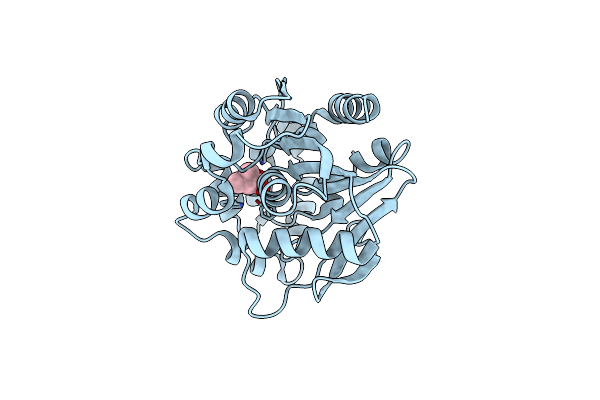 |
Crystal Structure Of A Meta-Cleavage Product Hydrolase (Cumd) Complexed With Isovalerate
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-09-14 Classification: HYDROLASE Ligands: IVA |
 |
Crystal Structure Of A Meta-Cleavage Product Hydrolase (Cumd) Complexed With (S)-2-Methylbutyrate
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2004-09-14 Classification: HYDROLASE Ligands: SMB |

