Search Count: 6
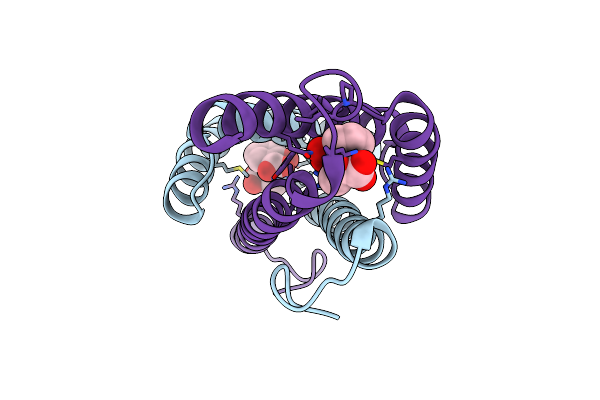 |
Structure Of The Isochorismate-Pyruvate Lyase From Pseudomonas Aerugionsa With Bound Salicylate And Pyruvate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2011-07-27 Classification: LYASE Ligands: SAL, PYR |
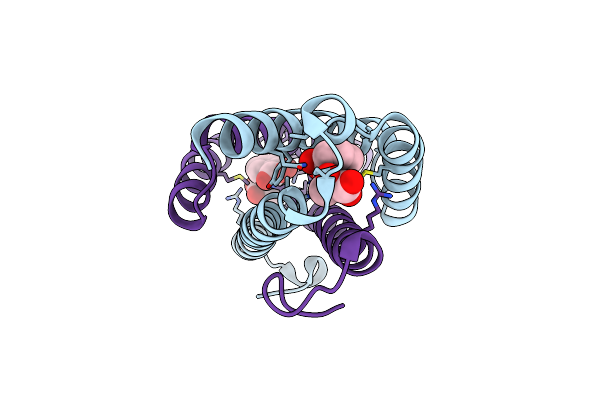 |
Salicylate And Pyruvate Bound Structure Of The Isochorismate-Pyruvate Lyase K42E Mutant From Pseudomonas Aerugionsa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2011-07-27 Classification: LYASE Ligands: SAL, PYR |
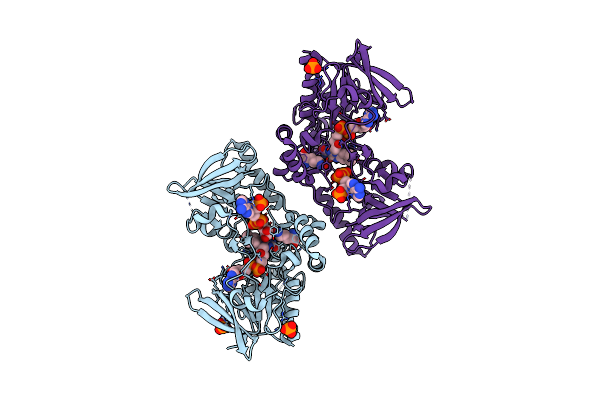 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-07-13 Classification: OXIDOREDUCTASE Ligands: FAD, PO4, ONH, NAP |
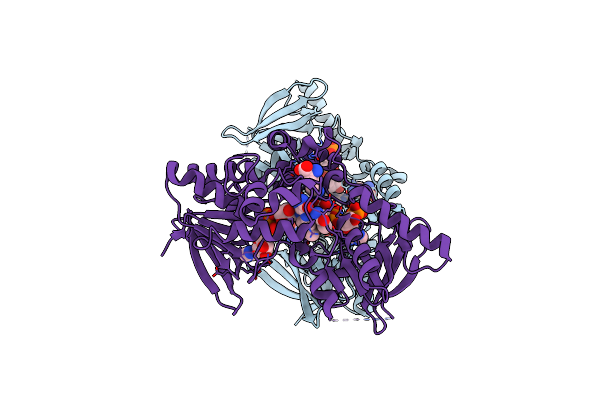 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2011-07-13 Classification: OXIDOREDUCTASE Ligands: FAD, ORN, NDP |
 |
Apo Structure Of Pseudomonas Aeruginosa Isochorismate-Pyruvate Lyase I87T Mutant
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2009-06-30 Classification: LYASE Ligands: CA |
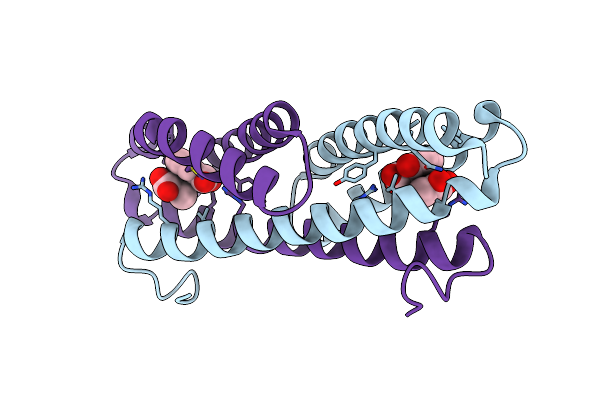 |
Crystal Structure Of Pseudomonas Aeruginosa Isochorismate-Pyruvate Lyase K42A Mutant In Complex With Salicylate And Pyruvate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-06-30 Classification: LYASE Ligands: SAL, PYR |

