Search Count: 47
 |
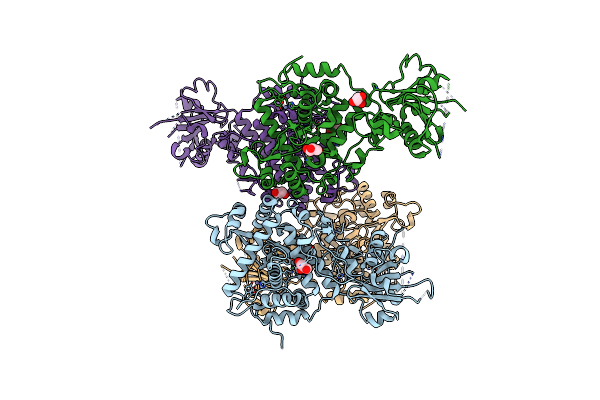
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: RNA BINDING PROTEIN Ligands: GOL |
 |
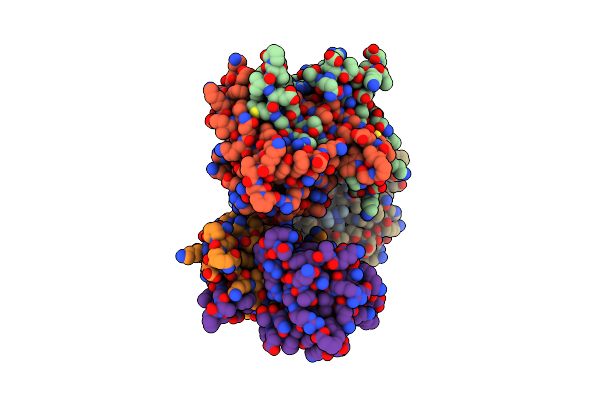
Three-Dimensional Structure Of Aip56, A Short-Trip Single Chain Ab Toxin From Photobacterium Damselae Subsp. Piscicida.
Organism: Photobacterium damselae subsp. piscicida
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2023-05-10 Classification: TOXIN Ligands: ZN, NI, GOL |
 |
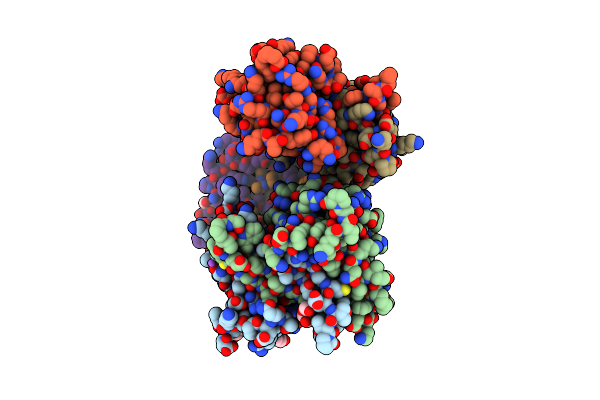
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-06-22 Classification: RNA BINDING PROTEIN, Viral protein Ligands: GOL, ACT |
 |
Crystal Structure Of Sars-Cov-2 Nucleocapsid Protein C-Terminal Domain Complexed With Chicoric Acid
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2022-06-22 Classification: RNA BINDING PROTEIN/Viral Protein Ligands: GKP, PEG, GOL, NA, CL |
 |
Crystal Structure Of Human Acetylcholinesterase In Complex With ((6-((2E,4E)-5-(Benzo[D][1,3]Dioxol-5-Yl)Penta-2,4-Dienamido)Hexyl)Triphenylphosphonium Bromide)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-06-09 Classification: HYDROLASE Ligands: NAG, QRH, SO4, BR, CL |
 |
Human Butyrylcholinesterase In Complex With ((6-((2E,4E)-5-(Benzo[D][1,3]Dioxol-5-Yl)Penta-2,4-Dienamido)Hexyl)Triphenylphosphonium Bromide)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-06-09 Classification: HYDROLASE Ligands: NAG, GOL, QRH, LEA, CL, SO4, BR |
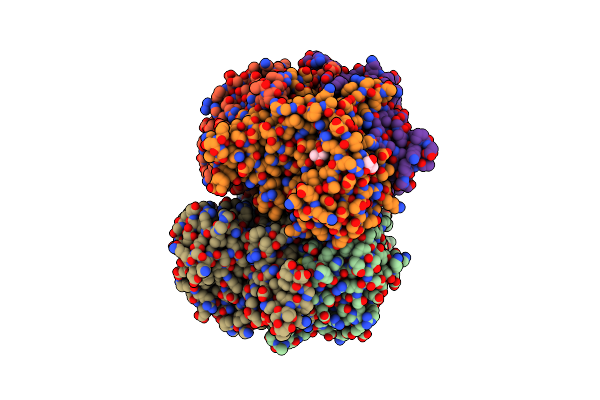 |
Crystal Structure Of Two-Domain Laccase Mutant R240H From Streptomyces Griseoflavus
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-05-05 Classification: OXIDOREDUCTASE Ligands: CU, GOL, EDO, OXY, MG |
 |
Crystal Structure Of Two-Domain Laccase Mutant R240A From Streptomyces Griseoflavus
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2021-05-05 Classification: OXIDOREDUCTASE Ligands: CU, GOL, OXY, PER |
 |
 |
Em Structure Of The Dna Wrapping In Bacterial Open Transcription Initiation Complex
Organism: Enterobacteria phage lambda, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2020-05-27 Classification: transcription/dna |
 |
Crystal Structure Of Two-Domain Laccase Mutant I170F From Streptomyces Griseoflavus
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-07-17 Classification: OXIDOREDUCTASE Ligands: CU, EDO, SO4, GOL, OXY |
 |
Crystal Structure Of Two-Domain Laccase Mutant I170A From Streptomyces Griseoflavus
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2019-07-17 Classification: OXIDOREDUCTASE Ligands: CU, GOL, SO4 |
 |
Crystal Structure Of Two-Domain Laccase From Streptomyces Griseoflavus Produced At 0.25 Mm Copper Sulfate In Growth Medium
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-07-17 Classification: OXIDOREDUCTASE Ligands: CU, PEO, EDO, GOL, SO4 |
 |
Crystal Structure Of The Gh51 Arabinofuranosidase From Xanthomonas Axonopodis Pv. Citri
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2019-02-20 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Two-Domain Laccase Mutant H165F From Streptomyces Griseoflavus With High Copper Ions Occupancy
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-01-30 Classification: OXIDOREDUCTASE Ligands: CU, PER, GOL, OXY |
 |
Crystal Structure Of Two-Domain Laccase Mutant H165A From Streptomyces Griseoflavus With High Copper Ions Occupancy
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2019-01-30 Classification: OXIDOREDUCTASE Ligands: CU, GOL, OXY |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-12-12 Classification: LIGASE, TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-12-12 Classification: LIGASE, TRANSFERASE |
 |
Crystal Structure Of The Gh2 Exo-Beta-Mannanase From Xanthomonas Axonopodis Pv. Citri
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-07-18 Classification: HYDROLASE/CARBOHYDRATE Ligands: GOL, PEG, ACT |
 |
Crystal Structure Of The Gh2 Exo-Beta-Mannanase From Xanthomonas Axonopodis Pv. Citri In Complex With Mannose
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2018-07-18 Classification: HYDROLASE/CARBOHYDRATE Ligands: BMA, ACT |

