Search Count: 24
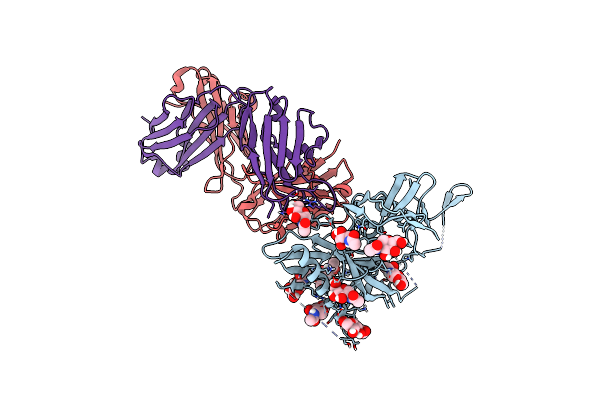 |
Crystal Structure Of Dh650 Fab From A Rhesus Macaque In Complex With Hiv-1 Gp120 Core
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-02 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
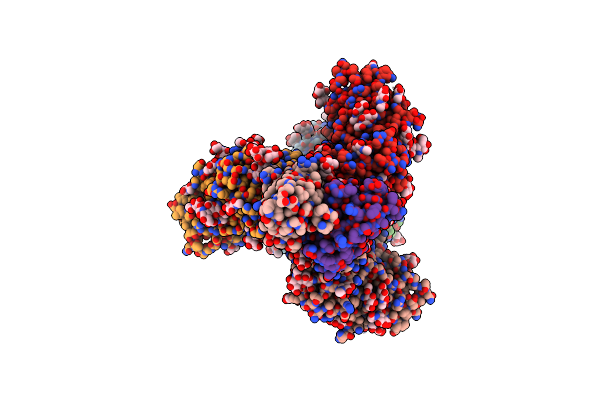 |
Cryo-Em Structure Of Shiv-Elicited Rha1.V2.01 In Complex With Hiv-1 Env Bg505 Ds-Sosip.664
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2020-10-07 Classification: IMMUNE SYSTEM/Viral Protein Ligands: NAG |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-05-03 Classification: HORMONE |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: 1PS, CL |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: 1PS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-06-25 Classification: ISOMERASE Ligands: PO4, GOL, K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2014-06-25 Classification: ISOMERASE Ligands: GOL |
 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:0.92 Å Release Date: 2014-06-25 Classification: ISOMERASE Ligands: GOL |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2013-09-04 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of N-Terminally Truncated Peroxiredoxin 4 From M. Musculus
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-09-04 Classification: OXIDOREDUCTASE Ligands: 1PE, ACY |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2013-09-04 Classification: ISOMERASE Ligands: PO4 |
 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-09-04 Classification: OXIDOREDUCTASE/ISOMERASE Ligands: K, GOL |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-03-23 Classification: HYDROLASE Ligands: CL, NA, ACT |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2011-03-23 Classification: HYDROLASE Ligands: ACT, NA, CL |
 |
High Resolution X-Ray Analysis Of Arg-Lysozyme Complex In The Presence Of 500 Mm Arg
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2011-03-23 Classification: HYDROLASE Ligands: ACT, ARG, NA, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-09-09 Classification: APOPTOSIS Ligands: ZN, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-09-09 Classification: APOPTOSIS Ligands: CA, 2PE, 12P |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-09-09 Classification: APOPTOSIS Ligands: MRD, PO4, NA |
 |
Crystal Structure Of Zn2+-Bound Form Of Des3-23Alg-2 Complexed With Alix Abs Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-09-09 Classification: APOPTOSIS Ligands: ZN, NA |
 |
Crystal Structures Of Ribonuclease Hi Active Site Mutants From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 1993-10-31 Classification: HYDROLASE(ENDORIBONUCLEASE) |

