Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Structure Of Sars-Cov-2 Ba.2.86 Spike Glycoprotein In Complex With Ace2 (2-Up State)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: VIRAL PROTEIN/PROTEIN BINDING Ligands: NAG |
 |
Structure Of Sars-Cov-2 Ba.2.86 Spike Glycoprotein In Complex With Ace2 (2-Up And 1-Down State)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: VIRAL PROTEIN/PROTEIN BINDING Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: VIRAL PROTEIN/PROTEIN BINDING Ligands: NAG |
 |
Structure Of Sars-Cov-2 Ba.2.86 Spike Rbd In Complex With Ace2 (Down State)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: VIRAL PROTEIN/PROTEIN BINDING Ligands: NAG |
 |
Structure Of Sars-Cov-2 Ba.2.86 Spike Glycoprotein In Complex With Ace2 (3-Up State)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: VIRAL PROTEIN/PROTEIN BINDING Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: VIRAL PROTEIN/PROTEIN BINDING Ligands: NAG |
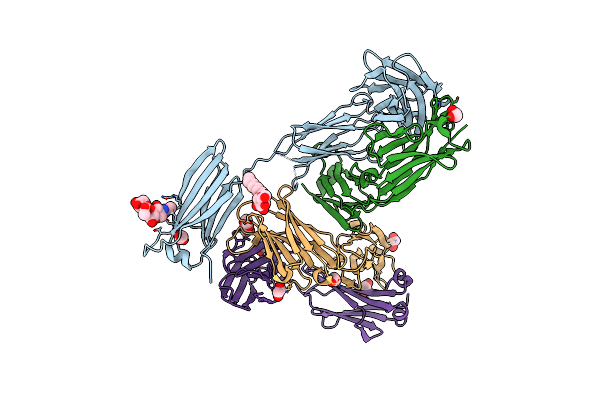 |
Crystal Structure Of Anti-Ige Antibody Hmk-12 Fab Complexed With Ige F(Ab')2
Organism: Rattus norvegicus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-07-31 Classification: IMMUNE SYSTEM Ligands: EDO, BOG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-12-14 Classification: METAL BINDING PROTEIN Ligands: EDO, ACT, PGE, SO4, ZN |
 |
Difructose Dianhydride I Synthase/Hydrolase (Alphaffase1) From Bifidobacterium Dentium, Ligand-Free Form
Organism: Bifidobacterium dentium
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: CA, MPD, TAR |
 |
Difructose Dianhydride I Synthase/Hydrolase (Alphaffase1) From Bifidobacterium Dentium In Complex With Beta-D-Arabinofuranose
Organism: Bifidobacterium dentium
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: CA, BXX |
 |
Difructose Dianhydride I Synthase/Hydrolase (Alphaffase1) From Bifidobacterium Dentium In Complex With Beta-D-Fructofuranose
Organism: Bifidobacterium dentium
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: CA, FRU |
 |
Organism: Sphingomonas sp
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-02-19 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Sphingomonas Sp. A1 Peroxidase Efeb Responsible For Import Of Iron
Organism: Sphingomonas sp. a1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-01-29 Classification: OXYGEN BINDING Ligands: HEM, OXY, PEG, EDO, PGE |
 |
Organism: Sphingomonas sp. a1
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2020-01-29 Classification: TRANSPORT PROTEIN Ligands: EDO, CIT |
 |
Organism: Sphingomonas sp. a1
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-10-04 Classification: STRUCTURAL PROTEIN Ligands: GOL, CU |