Search Count: 17
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-10-09 Classification: TRANSFERASE Ligands: PLS, A1L3N |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2024-10-09 Classification: TRANSFERASE Ligands: PGE |
 |
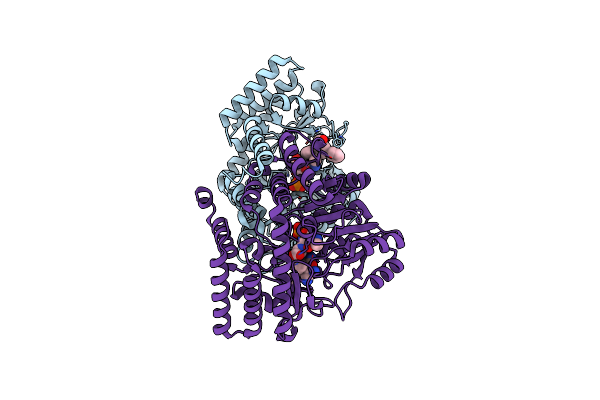
Complex Structure Of Serine Hydroxymethyltransferase From Enterococcus Faecium And Its Inhibitor
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: 5M5, PLP, CL |
 |
Crystal Structure Of E. Faecium Shmt In Complex With (+)-Shin-1 And Plp-Ser
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: 5M5, PLS |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: THH, PLG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2014-03-05 Classification: TRANSPORT PROTEIN Ligands: SO4, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-03-05 Classification: TRANSPORT PROTEIN Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-03-05 Classification: TRANSPORT PROTEIN Ligands: SO4 |
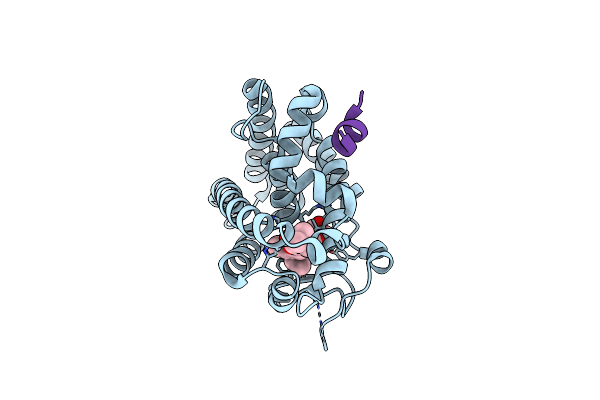 |
Crystal Structure Of The Rat Vitamin D Receptor Ligand Binding Domain Complexed With Yr335 And A Synthetic Peptide Containing The Nr2 Box Of Drip 205
Organism: Rattus norvegicus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2012-02-15 Classification: TRANSCRIPTION Ligands: YR4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2008-08-05 Classification: PROTEIN TRANSPORT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-08-05 Classification: PROTEIN TRANSPORT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2007-09-04 Classification: SIGNALING PROTEIN ACTIVATOR Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-09-04 Classification: SIGNALING PROTEIN ACTIVATOR Ligands: P6G, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2003-07-15 Classification: PROTEIN TRANSPORT Ligands: ZN, MG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2003-07-15 Classification: PROTEIN TRANSPORT Ligands: SO4 |
 |
Crystal Structure Of The Outer Membrane Lipoprotein Receptor Lolb Complexed With Pegmme2000
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2003-07-15 Classification: PROTEIN TRANSPORT Ligands: SO4, PG5 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2003-07-15 Classification: PROTEIN TRANSPORT |

