Search Count: 13
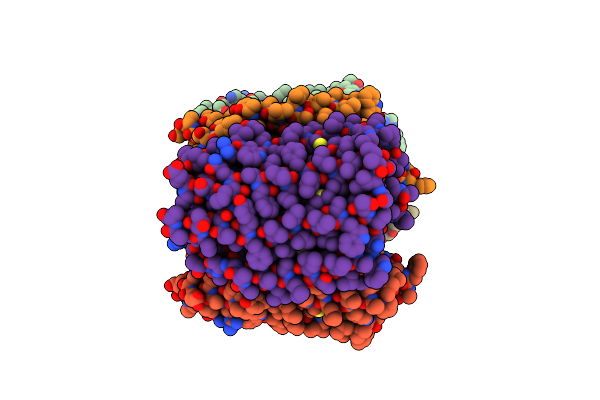 |
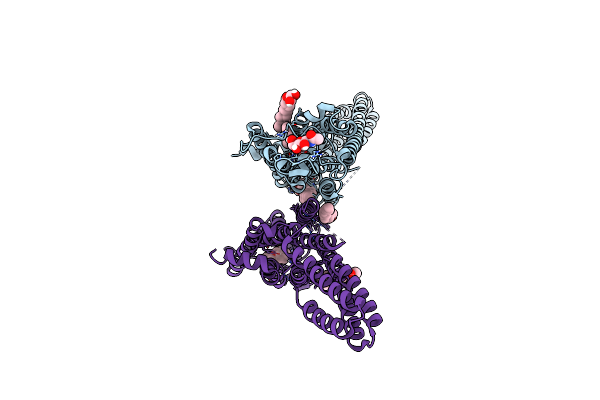
Crystal Structure Of The Microbial Rhodopsin From Sphingomonas Paucimobilis (Spar)
Organism: Sphingomonas paucimobilis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-05-03 Classification: MEMBRANE PROTEIN Ligands: LFA |
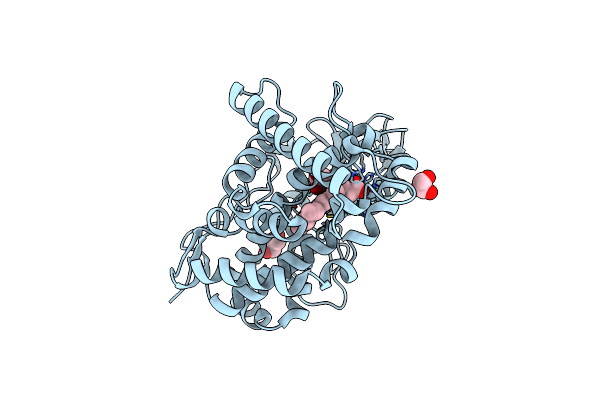 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: HEM, CL, GOL |
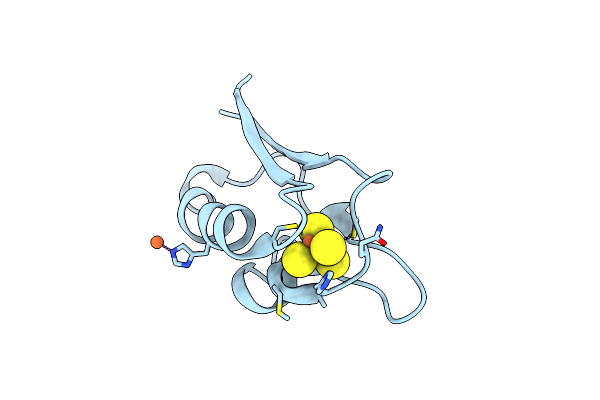 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-02-15 Classification: ELECTRON TRANSPORT Ligands: F3S, FE |
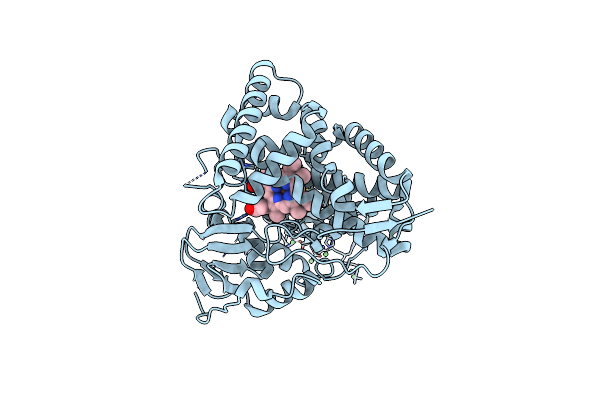 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: F3S, HEM, NI |
 |
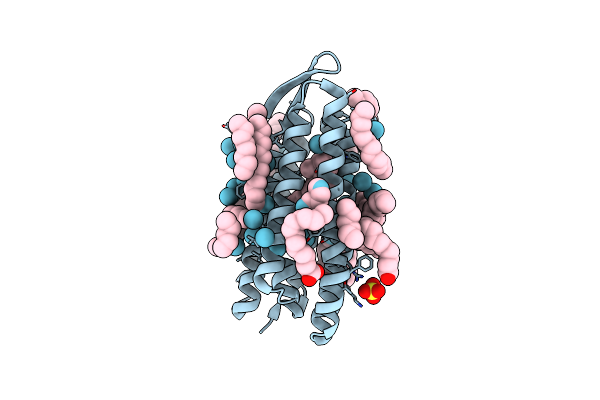
Xfel Crystal Structure Of The Human Sphingosine 1 Phosphate Receptor 5 In Complex With Ono-5430608
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-08-10 Classification: MEMBRANE PROTEIN Ligands: NAG, B2O, OLC, OLA |
 |
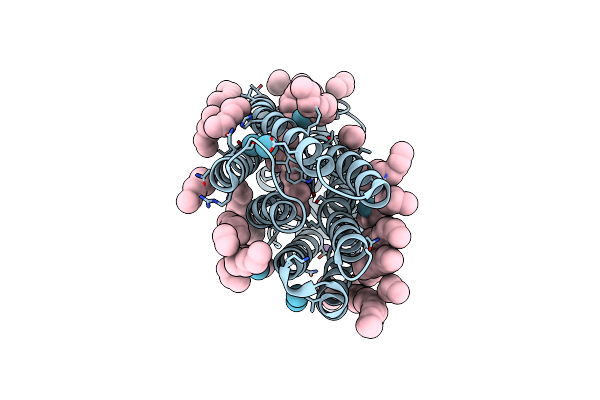
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-04-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA, OLA, HEX, SO4, KR |
 |
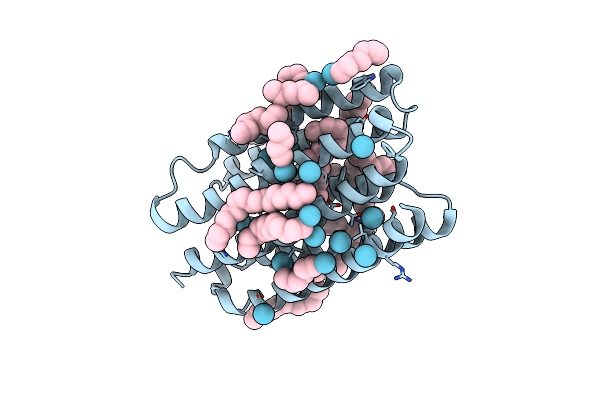
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-04-27 Classification: MEMBRANE PROTEIN Ligands: LFA, HEX, NA, KR, RET |
 |
Organism: Candidatus actinomarina minuta
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-04-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA, HEX, KR |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-04-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA, OLC, OLA, HEX, SO4, AR |
 |
Crystal Structure Of A Light-Driven Proton Pump Lr (Mac) From Leptosphaeria Maculans
Organism: Leptosphaeria maculans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-07-07 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA |
 |
Dynamic Complex Between All-D-Enantiomeric Peptide D3 With Wild-Type Amyloid Precursor Protein 672-726 Fragment (Amyloid Beta 1-55)
Organism: Homo sapiens, Synthetic construct
Method: SOLUTION NMR Release Date: 2021-01-13 Classification: PROTEIN BINDING |
 |
Dynamic Complex Between All-D-Enantiomeric Peptide D3 With L723P Mutant Of Amyloid Precursor Protein (App) 672-726 Fragment (Amyloid Beta 1-55)
Organism: Homo sapiens, Synthetic construct
Method: SOLUTION NMR Release Date: 2021-01-13 Classification: PROTEIN BINDING |
 |
Crystal Structure Of The C14 Ring Of The F1Fo Atp Synthase From Spinach Chloroplast
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-12-25 Classification: MEMBRANE PROTEIN Ligands: LFA, OLC |

