Search Count: 42
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: ELECTRON TRANSPORT |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: ELECTRON TRANSPORT |
 |
Organism: Sulfolobus shibatae b12
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-11-21 Classification: HYDROLASE Ligands: MG, GOL |
 |
Organism: Chromohalobacter sp. 560
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-03-04 Classification: HYDROLASE |
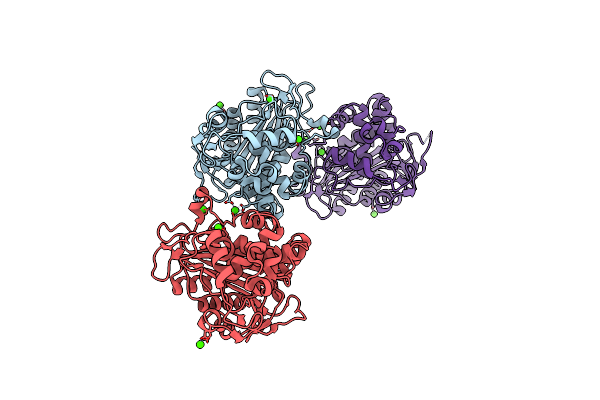 |
N288Q-N321Q Mutant Beta-Lactamase Derived From Chromohalobacter Sp.560 (Without Soaking)
Organism: Chromohalobacter sp. 560
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: CA, CL |
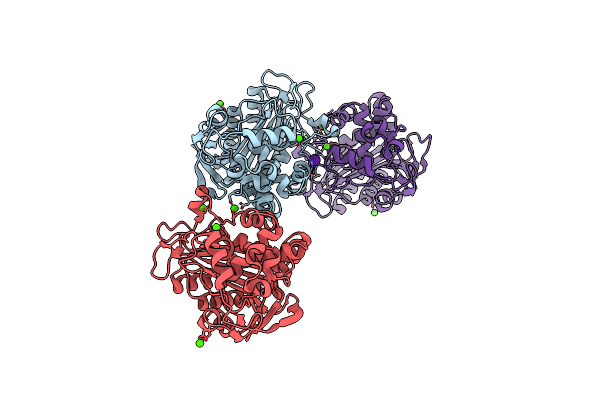 |
N288Q-N321Q Mutant Beta-Lactamase Derived From Chromohalobacter Sp.560 (Condition-1A)
Organism: Chromohalobacter sp. 560
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: CA, CS, CL |
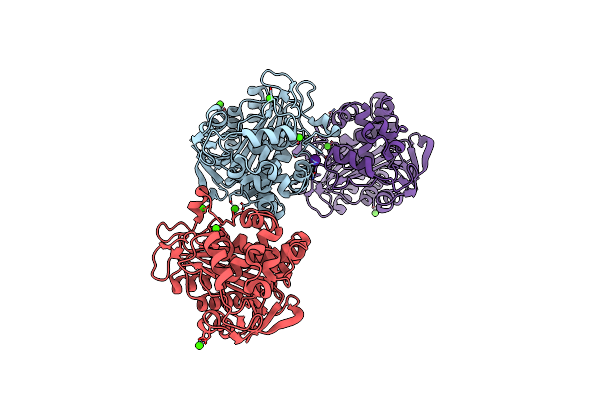 |
N288Q-N321Q Mutant Beta-Lactamase Derived From Chromohalobacter Sp.560 (Condition-1B)
Organism: Chromohalobacter sp. 560
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: CS, CA |
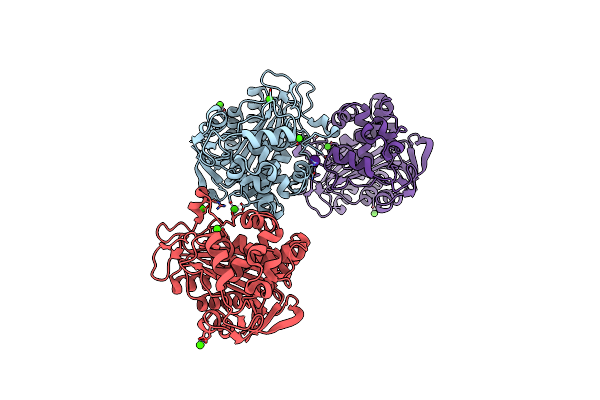 |
N288Q-N321Q Mutant Beta-Lactamase Derived From Chromohalobacter Sp.560 (Condition-1C)
Organism: Chromohalobacter sp. 560
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: CS, CA |
 |
N288Q-N321Q Mutant Beta-Lactamase Derived From Chromohalobacter Sp.560 (Condition-2A)
Organism: Chromohalobacter sp. 560
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: SR, CL |
 |
N288Q-N321Q Mutant Beta-Lactamase Derived From Chromohalobacter Sp.560 (Condition-2B)
Organism: Chromohalobacter sp. 560
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: SR, CL, CA |
 |
Crystal Structure Of Catalytic Domain Of Chitinase From Ralstonia Sp. A-471
Organism: Ralstonia
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Catalytic Domain Of Chitinase From Ralstonia Sp. A-471 In Complex With Disaccharide
Organism: Ralstonia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-05-15 Classification: HYDROLASE |
 |
Crystal Structure Of Catalytic Domain Of Chitinase From Ralstonia Sp. A-471 (E141Q) In Complex With Tetrasaccharide
Organism: Ralstonia
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2013-05-15 Classification: HYDROLASE |
 |
Crystal Structure Of Catalytic Domain Of Chitinase From Ralstonia Sp. A-471 (E162Q)
Organism: Ralstonia
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: EPE, TRS |
 |
Crystal Structure Of Catalytic Domain Of Chitinase From Ralstonia Sp. A-471 (E162Q) In Complex With Disaccharide
Organism: Ralstonia
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-05-15 Classification: HYDROLASE |
 |
Organism: Halomonas
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-07-11 Classification: TRANSFERASE |
 |
Organism: Halomonas
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-07-11 Classification: TRANSFERASE |
 |
Organism: Halomonas
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-07-11 Classification: TRANSFERASE |
 |
Organism: Halomonas
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-07-11 Classification: TRANSFERASE |
 |
Crystal Structure Of Glycosyltrehalose Trehalohydrolase (Gthase) From Sulfolobus Solfataricus Km1
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2012-06-20 Classification: HYDROLASE Ligands: FLC, GOL |

