Search Count: 16
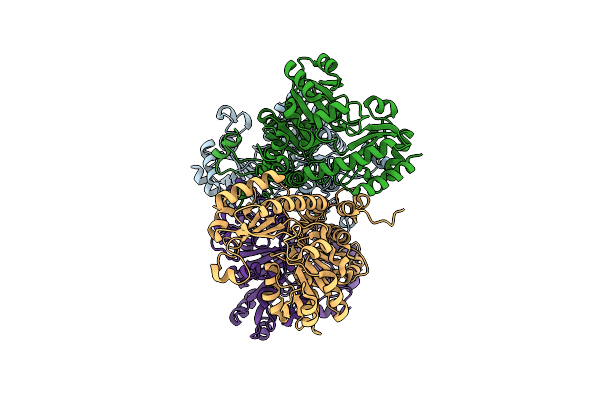 |
Crystal Structure Of Pseudomonas Putida Methionine Gamma-Lyase Q349S Mutant Ligand-Free Form.
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-04-20 Classification: LYASE |
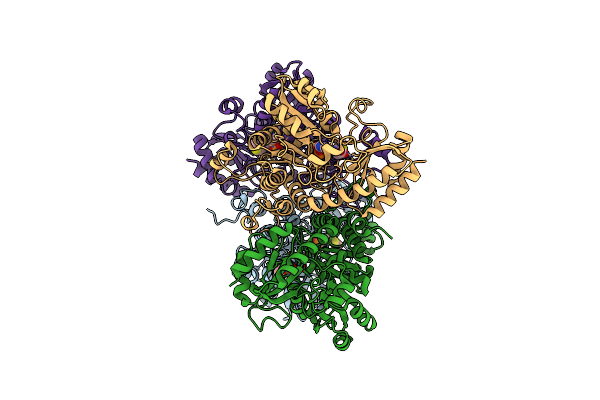 |
Crystal Structure Of Pseudomonas Putida Methionine Gamma-Lyase Q349S Mutant With L-Methionine Intermediates
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-04-20 Classification: LYASE Ligands: 3LM, MET |
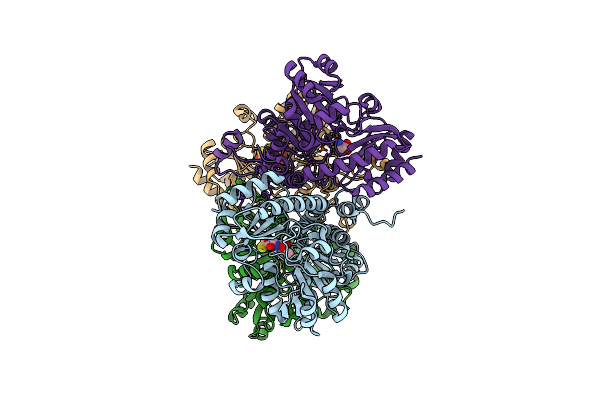 |
Crystal Structure Of Pseudomonas Putida Methionine Gamma-Lyase Q349S Mutant With L-Homocysteine Intermediates
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Release Date: 2022-04-20 Classification: LYASE Ligands: 7XF, HCS |
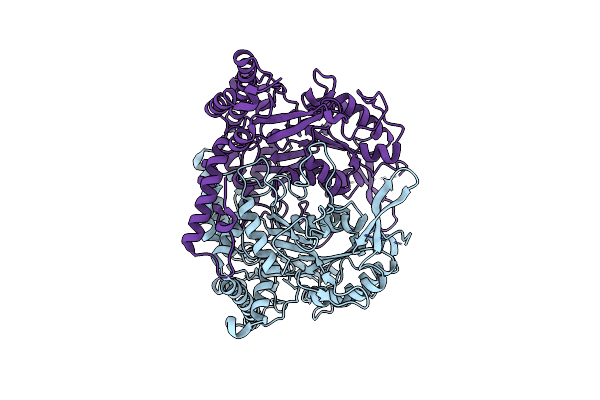 |
Crystal Structure Of L-Methionine Decarboxylase From Streptomyces Sp.590 (Internal Aldimine Form).
Organism: Streptomyces sp. 590 ki-2014
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-01-27 Classification: LYASE |
 |
Crystal Structure Of L-Methionine Decarboxylase Q64A Mutant From Streptomyces Sp.590 In Complexed With L- Methionine Methyl Ester (Geminal Diamine Form).
Organism: Streptomyces sp. 590 ki-2014
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-01-27 Classification: LYASE Ligands: G03 |
 |
Crystal Structure Of L-Methionine Decarboxylase From Streptomyces Sp.590 In Complexed With L- Methionine Methyl Ester (External Aldimine Form).
Organism: Streptomyces sp. 590 ki-2014
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2021-01-27 Classification: LYASE Ligands: G06 |
 |
Crystal Structure Of L-Methionine Decarboxylase From Streptomyces Sp.590 In Complexed With 3-Methlythiopropylamine (External Aldimine Form).
Organism: Streptomyces sp. 590 ki-2014
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2021-01-27 Classification: LYASE Ligands: G0C |
 |
Crystal Structure Of L-Methionine Decarboxylase From Streptomyces Sp.590 In Complexed With 3-Methlythiopropylamine (Geminal Diamine Form).
Organism: Streptomyces sp. 590 ki-2014
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-01-27 Classification: LYASE Ligands: G0F |
 |
Structure-Based Design Of Ask1 Inhibitors As Potential First-In-Class Agents For Heart Failure
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2017-06-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: SO4, 8GV |
 |
Structure-Based Design Of Ask1 Inhibitors As Potential First-In-Class Agents For Heart Failure
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-06-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 8GY |
 |
Structure-Based Design Of Ask1 Inhibitors As Potential First-In-Class Agents For Heart Failure
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2017-06-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 8GS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-03-30 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: SO4, 03P |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2011-03-30 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 03Q |
 |
Organism: Geobacillus sp. mo-1
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2010-09-01 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, 3A2, ACT |
 |
Organism: Geobacillus sp. mo-1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-08-25 Classification: HYDROLASE Ligands: ZN, ACT |
 |
Organism: Geobacillus sp. mo-1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-08-25 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 3A1, ZN, ACT |

