Search Count: 31
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: CYTOKINE Ligands: SO4, GOL, IPA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: CYTOKINE Ligands: SO4, GOL, IPA |
 |
Macrophage Migration Inhibitory Factor S61H/Y100H Mutant Complexed With Three Zinc Ions (Zn3-Mif(S61H/Y100H)-L)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: CYTOKINE Ligands: ZN, CO3, SO4, GOL, IPA |
 |
Macrophage Migration Inhibitory Factor S61H/Y100H Mutant Complexed With Three Zinc Ions (Zn3-Mif(S61H/Y100H))
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: CYTOKINE Ligands: ZN, CO3, SO4, GOL, IPA |
 |
Macrophage Migration Inhibitory Factor Y100H Mutant Complexed With Three Zinc Ions (Zn3-Mif(Y100H))
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: CYTOKINE Ligands: ZN, CL, CO3, SO4, GOL, IPA |
 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Substrate 8-Oxoguanine Dna At Ph 8.0 Under 277 K
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: PEG, GOL, MG, NA |
 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 277 K For 24 Hourss
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, MG, GOL |
 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 277 K For 2.5 Hours
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, MG, GOL |
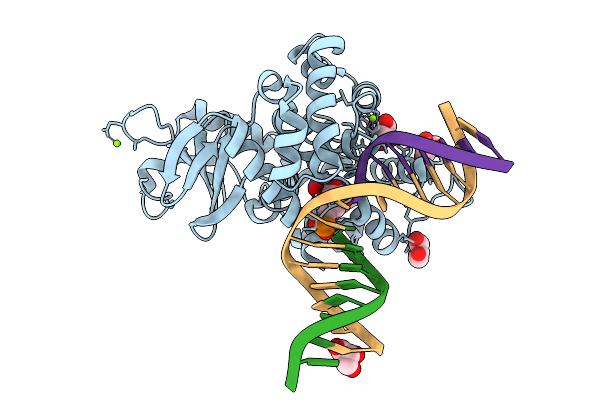 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 298 K For 3 Weeks
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, GOL, MG, NA |
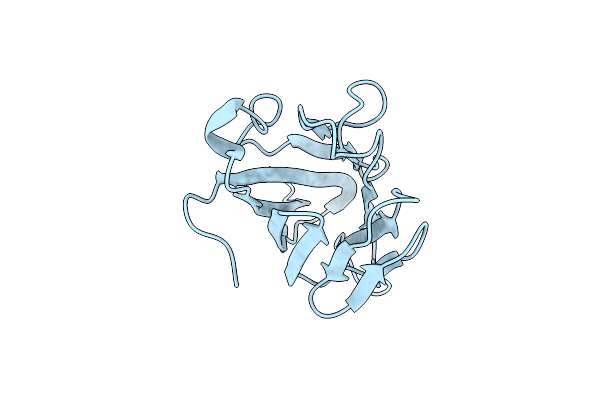 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-04-01 Classification: SIGNALING PROTEIN |
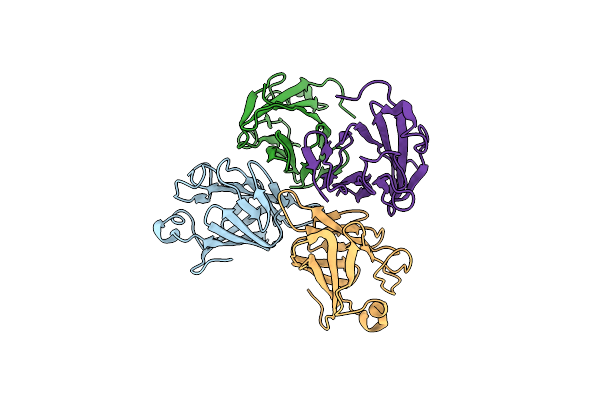 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2020-04-01 Classification: SIGNALING PROTEIN |
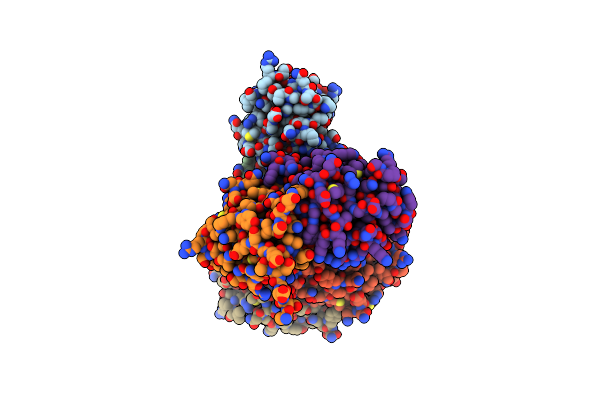 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-04-01 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of The N-Terminal Domain Of Htaa From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-11-20 Classification: HEME-BINDING PROTEIN Ligands: HEM |
 |
Crystal Structure Of The C-Terminal Domain Of Htaa From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-11-20 Classification: HEME-BINDING PROTEIN Ligands: HEM |
 |
Crystal Structure Of Heme Binding Protein Htab From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-11-20 Classification: HEME-BINDING PROTEIN Ligands: HEM, CA |
 |
Crystal Structure Of The H434A Variant Of The C-Terminal Domain Of Htaa From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-11-20 Classification: HEME-BINDING PROTEIN Ligands: HEM, NA |
 |
Crystal Structure Of The Domain-Swapped Dimer H434A Variant Of The C-Terminal Domain Of Htaa From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2019-11-20 Classification: HEME-BINDING PROTEIN Ligands: IPA |
 |
Organism: Kluyveromyces lactis (strain atcc 8585 / cbs 2359 / dsm 70799 / nbrc 1267 / nrrl y-1140 / wm37)
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2017-11-08 Classification: LIPID BINDING PROTEIN |
 |
Organism: Kluyveromyces lactis (strain atcc 8585 / cbs 2359 / dsm 70799 / nbrc 1267 / nrrl y-1140 / wm37)
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2017-11-08 Classification: LIPID BINDING PROTEIN |
 |
Organism: Kluyveromyces lactis (strain atcc 8585 / cbs 2359 / dsm 70799 / nbrc 1267 / nrrl y-1140 / wm37)
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2017-11-08 Classification: LIPID BINDING PROTEIN Ligands: 6OU, K |

