Search Count: 22
 |
Organism: Synthetic construct, Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE/RNA Ligands: ZN, GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: GOL, ZN |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FXU, ZN, MG, CL |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FXX, MPO, ZN |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FY6, ZN, CL |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FY9, ZN |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FYF, ZN, MG |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FYL, PO4, EDO, ZN |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: MPB, ZN |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FYR, DMS, ZN |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FZ0, ZN |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FZ3, DMS, ZN |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FZ6, ZN |
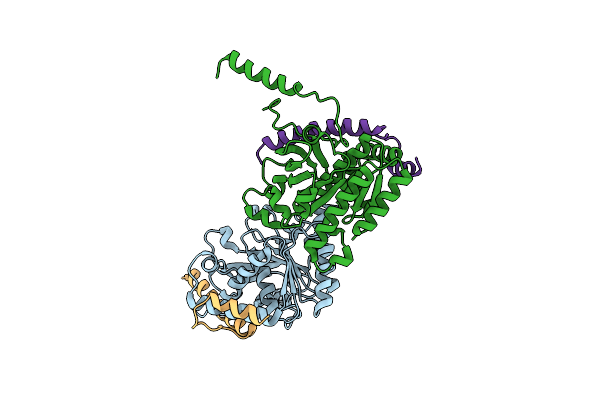 |
Crystal Structure Of The Complex Of The Interaction Domains Of E. Coli Dnab Helicase And Dnac Helicase Loader
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2019-11-20 Classification: HYDROLASE |
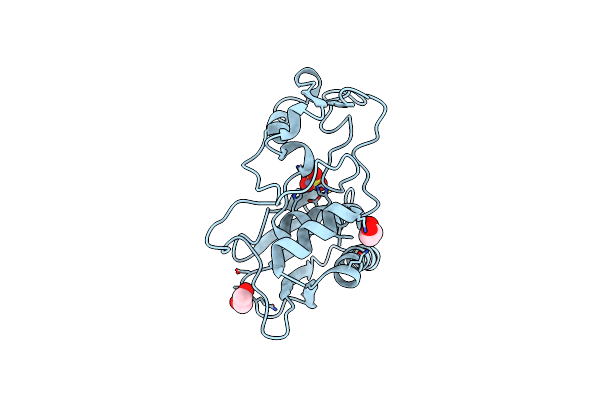 |
Crystal Structure Of The Hatching Enzyme Zhe1 From The Zebrafish Danio Rerio
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2010-09-08 Classification: HYDROLASE Ligands: SO4, ZN, EDO |
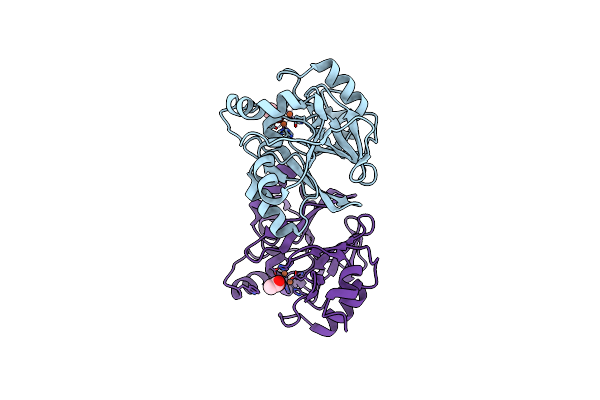 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-01-05 Classification: HYDROLASE Ligands: FE, ACT |
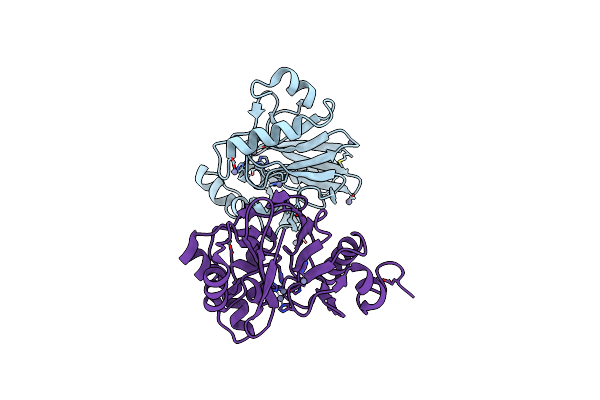 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-10-06 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-09-12 Classification: TRANSCRIPTION |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-07-26 Classification: HYDROLASE Ligands: ZN, SER |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2005-07-26 Classification: HYDROLASE Ligands: ZN |

