Search Count: 6
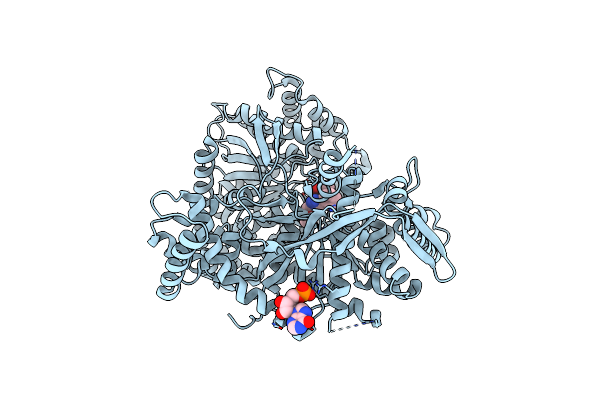 |
Crystal Structure Of 1-(Beta-D-Glucopyranosyl)-4-Substituted-1,2,3-Triazoles In Complex With Glycogen Phosphorylase
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2010-02-16 Classification: TRANSFERASE Ligands: KOT, DMS, IMP |
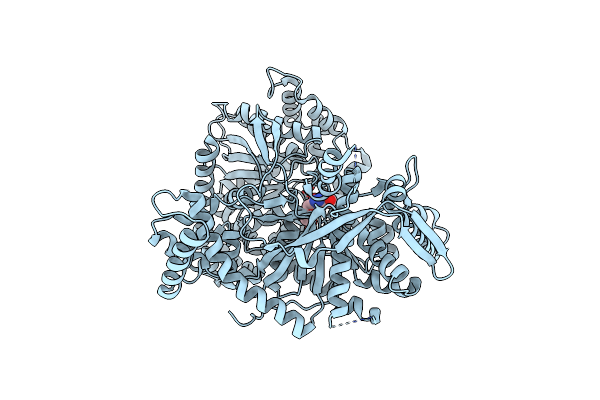 |
Crystal Structure Of 1-(Beta-D-Glucopyranosyl)-4-Substituted-1,2,3-Triazole
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-02-16 Classification: TRANSFERASE Ligands: RUG |
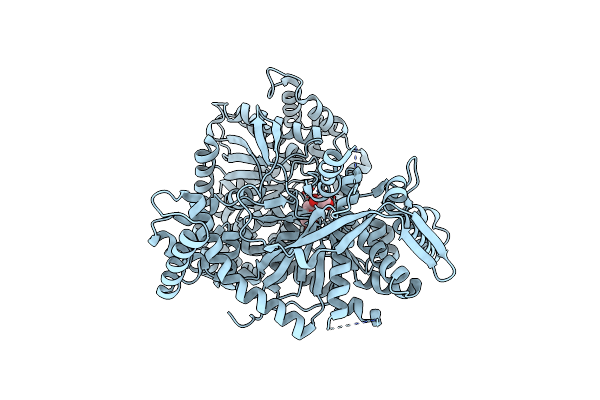 |
Crystal Structure Of 1-(Beta-D-Glucopyranosyl)-4-Substituted-1,2,3-Triazoles In Complex With Glycogen Phosphorylase
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2010-02-16 Classification: TRANSFERASE Ligands: 9GP |
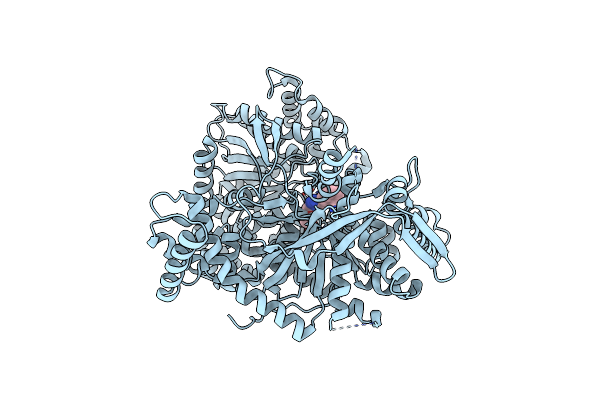 |
Crystal Structure Of 1-(Beta-D-Glucopyranosyl)-4-Substituted-1,2,3-Triazole
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-02-16 Classification: TRANSFERASE Ligands: SKY |
 |
Crystal Structure Of 1-(Beta-D-Glucopyranosyl)-4-Substituted-1,2,3-Triazoles In Complex With Glycogen Phosphorylase
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-02-16 Classification: TRANSFERASE Ligands: LEW |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-02-16 Classification: TRANSFERASE Ligands: OAK |

