Search Count: 49
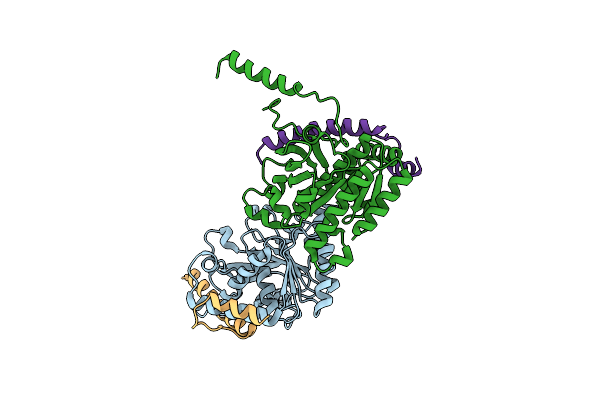 |
Crystal Structure Of The Complex Of The Interaction Domains Of E. Coli Dnab Helicase And Dnac Helicase Loader
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2019-11-20 Classification: HYDROLASE |
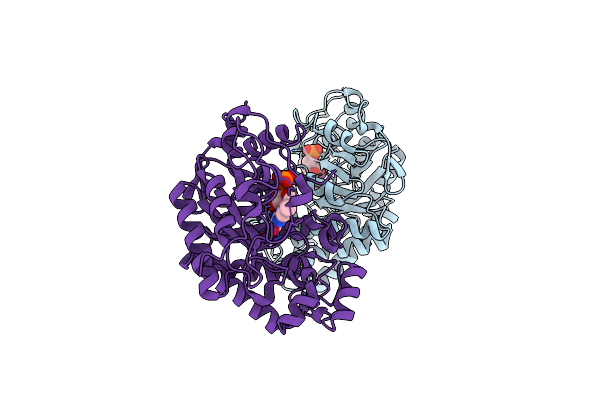 |
Organism: Pichia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-06-26 Classification: OXIDOREDUCTASE Ligands: FMN |
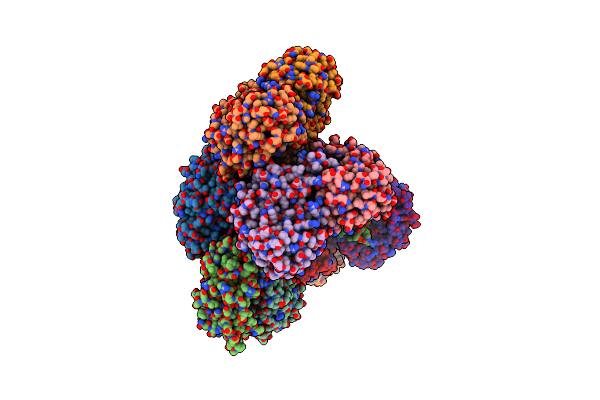 |
Crystal Structure Of The G136F Mutant Of The First R-Stereoselective -Transaminase Identified From Arthrobacter Sp. Knk168 (Ferm-Bp-5228)
Organism: Arthrobacter sp. knk168
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2015-08-19 Classification: TRANSFERASE Ligands: PLP |
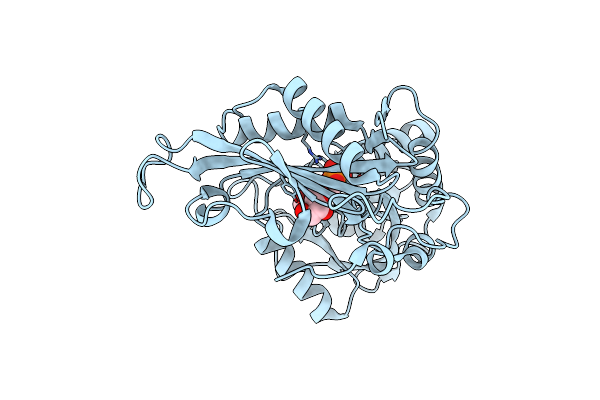 |
Crystal Structure Of The First R-Stereoselective -Transaminase Identified From Arthrobacter Sp. Knk168 (Ferm-Bp-5228)
Organism: Arthrobacter sp. knk168
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2015-08-12 Classification: TRANSFERASE Ligands: PLP, GOL |
 |
Crystal Structure Of An Engineered Sitagliptin-Producing Transaminase, Ata-117-Rd11
Organism: Arthrobacter sp. knk168
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-08-12 Classification: TRANSFERASE Ligands: PLP |
 |
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2015-02-25 Classification: OXIDOREDUCTASE |
 |
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-02-25 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Organism: Kluyveromyces marxianus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-02-11 Classification: FLAVOPROTEIN Ligands: FMN |
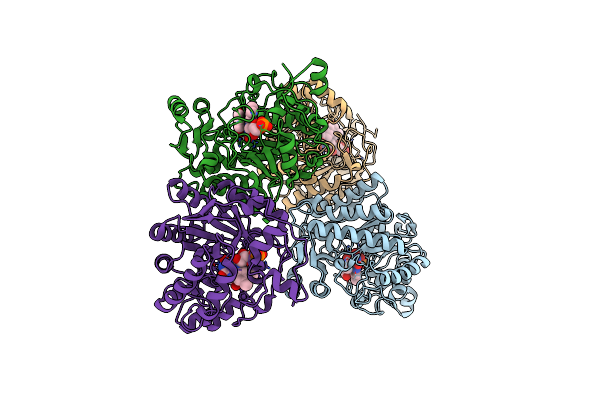 |
Crystal Structure Of Old Yellow Enzyme From Candida Macedoniensis Aku4588 Complexed With P-Hydroxybenzaldehyde
Organism: Kluyveromyces marxianus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-02-11 Classification: FLAVOPROTEIN Ligands: FMN, HBA |
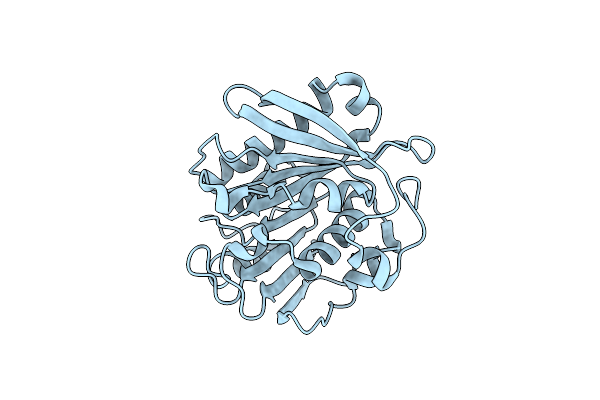 |
Crystal Structure Of Pet-Degrading Cutinase Cut190 S226P Mutant In Ca(2+)-Free State
Organism: Saccharomonospora viridis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2014-12-24 Classification: HYDROLASE |
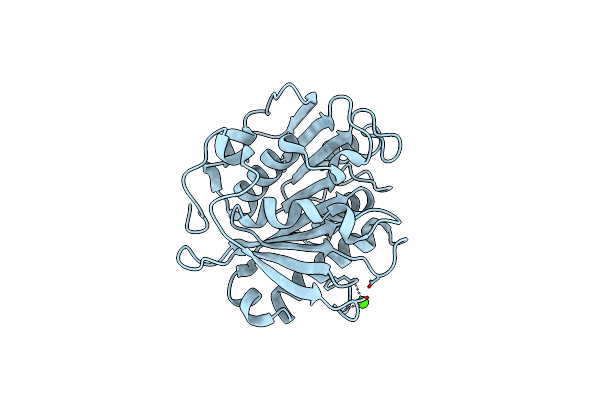 |
Crystal Structure Of Pet-Degrading Cutinase Cut190 S226P Mutant In Ca(2+)-Bound State At 1.75 Angstrom Resolution
Organism: Saccharomonospora viridis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-12-24 Classification: HYDROLASE Ligands: CA, CL |
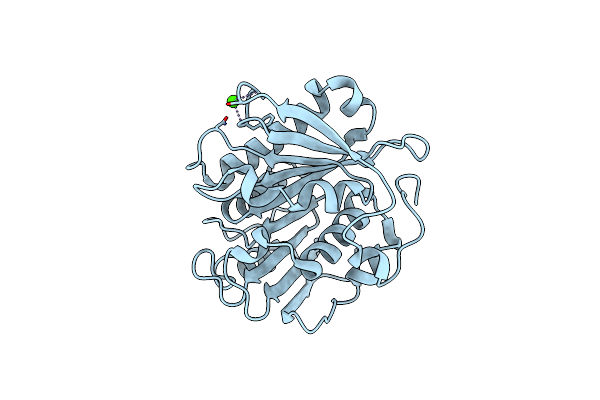 |
Crystal Structure Of Pet-Degrading Cutinase Cut190 S226P Mutant In Ca(2+)-Bound State At 2.35 Angstrom Resolution
Organism: Saccharomonospora viridis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2014-12-24 Classification: HYDROLASE Ligands: CA, CL |
 |
Crystal Structure Of Low-Specificity L-Threonine Aldolase From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2014-12-17 Classification: LYASE Ligands: PLG |
 |
Crystal Structure Of The Novel Haloalkane Dehalogenase Data From Agrobacterium Tumefaciens C58
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-07-23 Classification: HYDROLASE Ligands: NHE, GOL |
 |
Crystal Structure Of Y109W Mutant Haloalkane Dehalogenase Data From Agrobacterium Tumefaciens C58
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-07-23 Classification: HYDROLASE Ligands: NHE |
 |
Organism: Aeromonas jandaei
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-07-09 Classification: LYASE Ligands: PLG, GLY |
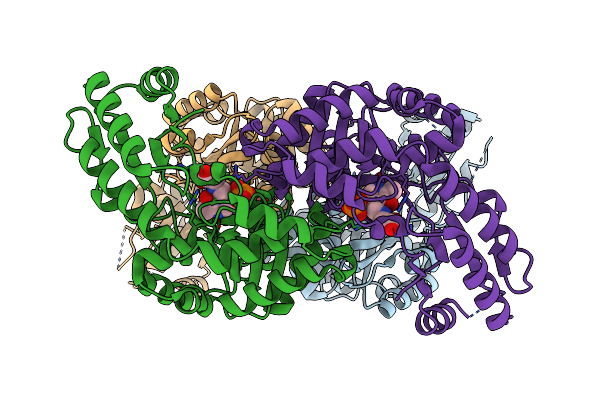 |
Organism: Aeromonas jandaei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-07-09 Classification: LYASE Ligands: PLG |
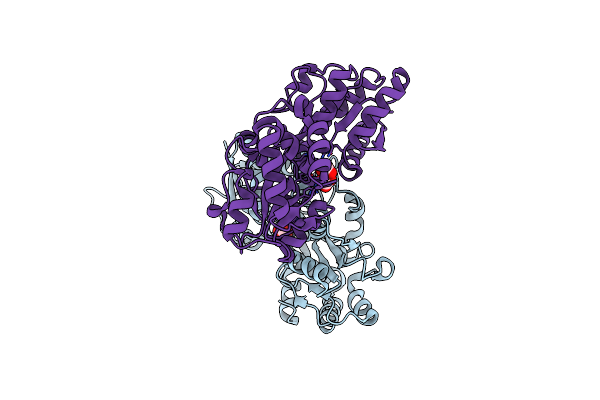 |
X-Ray Structure Of Fe(Iii)-Bicarbonates-Ttfbpa, A Ferric Ion-Binding Protein From Thermus Thermophilus Hb8
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-05-21 Classification: METAL BINDING PROTEIN Ligands: FE, BCT |
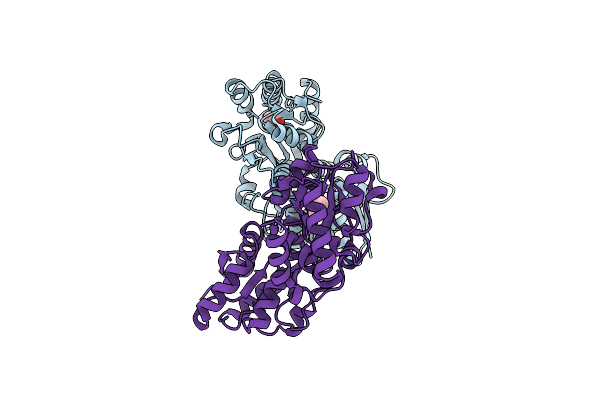 |
X-Ray Structure Of Apo-Ttfbpa, A Ferric Ion-Binding Protein From Thermus Thermophilus Hb8
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-05-21 Classification: METAL BINDING PROTEIN Ligands: IPA |
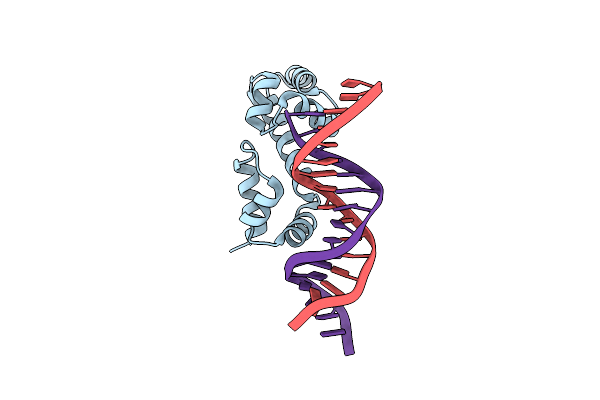 |
Crystal Structure Of The Dna-Binding Domain Of Adpa, The Global Transcriptional Factor, In Complex With A Target Dna
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2013-09-18 Classification: TRANSCRIPTION ACTIVATOR/DNA |

