Search Count: 43
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: ELECTRON MICROSCOPY Release Date: 2020-10-28 Classification: UNKNOWN FUNCTION Ligands: LMN |
 |
Organism: Enterobacteria phage rb59, Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2019-03-20 Classification: LIPID TRANSPORT |
 |
Organism: Enterobacteria phage rb59, Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-03-20 Classification: LIPID TRANSPORT Ligands: PEE |
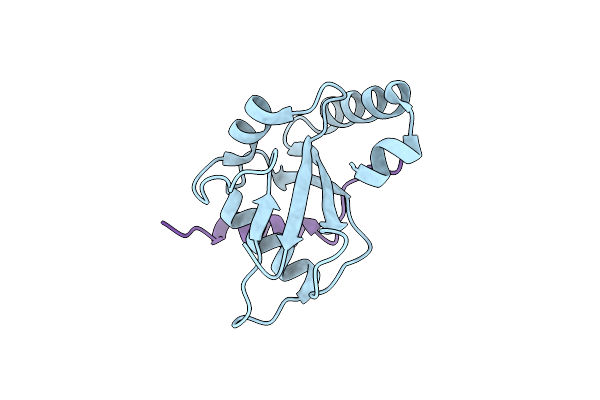 |
Crystal Structure Of Fission Yeast Atg8 Complexed With The Helical Aim Of Hfl1.
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-12-12 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of Budding Yeast Atg8 Complexed With The Helical Aim Of Hfl1.
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae (strain yjm789)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2018-12-12 Classification: MEMBRANE PROTEIN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-03-28 Classification: PROTEIN TRANSPORT/METAL BINDING PROTEIN Ligands: ZN, ATP, MG |
 |
Organism: Lachancea thermotolerans (strain atcc 56472 / cbs 6340 / nrrl y-8284), Lachancea thermotolerans
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2016-08-03 Classification: PROTEIN TRANSPORT |
 |
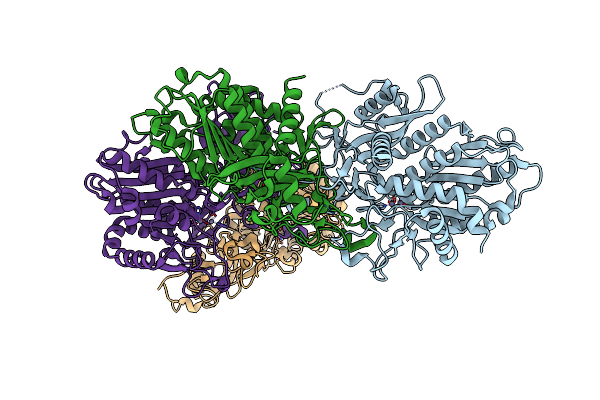
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2016-06-29 Classification: PROTEIN TRANSPORT |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2016-06-29 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-06-29 Classification: HYDROLASE Ligands: ZN, CAC |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2016-06-29 Classification: HYDROLASE |
 |
Organism: Kluyveromyces marxianus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-05-07 Classification: PROTEIN TRANSPORT |
 |
Organism: Lachancea thermotolerans
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2014-05-07 Classification: PROTEIN TRANSPORT |
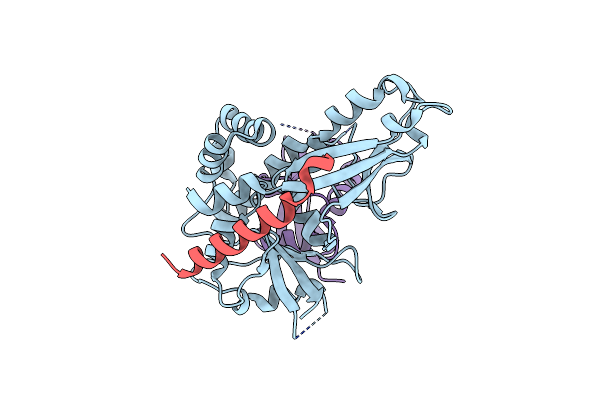 |
Crystal Structure Of Saccharomyces Cerevisiae Atg12-Atg5 Conjugate Bound To The N-Terminal Domain Of Atg16
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-12-26 Classification: LIGASE |
 |
Organism: Kluyveromyces marxianus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-11-14 Classification: LIGASE |
 |
Organism: Kluyveromyces marxianus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2012-11-14 Classification: LIGASE |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2012-11-14 Classification: LIGASE |
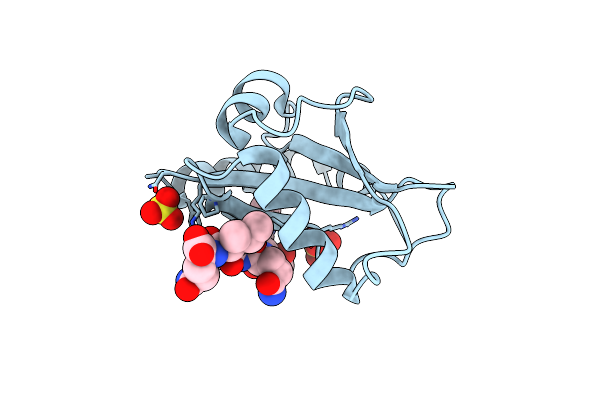 |
Crystal Structure Of Saccharomyces Cerevisiae Atg8 Complexed With Atg32 Aim
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-10-03 Classification: PROTEIN TRANSPORT Ligands: SO4 |
 |
Organism: Kluyveromyces marxianus
Method: SOLUTION NMR Release Date: 2012-08-01 Classification: PROTEIN TRANSPORT |
 |
Organism: Kluyveromyces marxianus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-08-01 Classification: PROTEIN TRANSPORT Ligands: EPE, SO4 |

