Search Count: 13
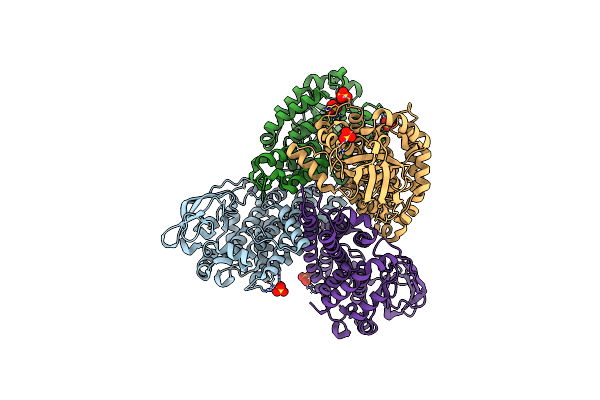 |
Crystal Structure Of Dibenzothiophene Monooxygenase (Tdsc) From Paenibacillus Sp. A11-2
Organism: Paenibacillus sp. a11-2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-08-09 Classification: OXIDOREDUCTASE Ligands: SO4 |
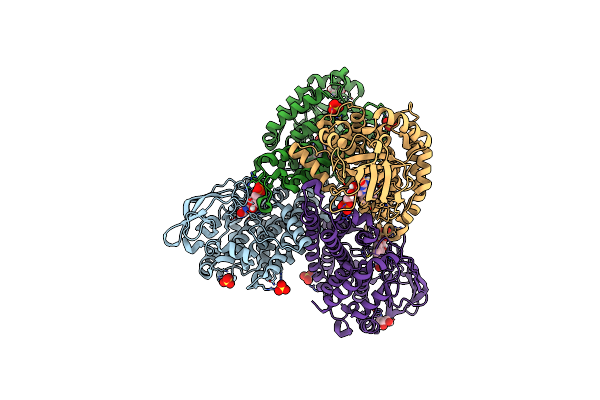 |
Organism: Paenibacillus sp. a11-2
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2017-08-09 Classification: OXIDOREDUCTASE Ligands: FMN, SO4, GOL |
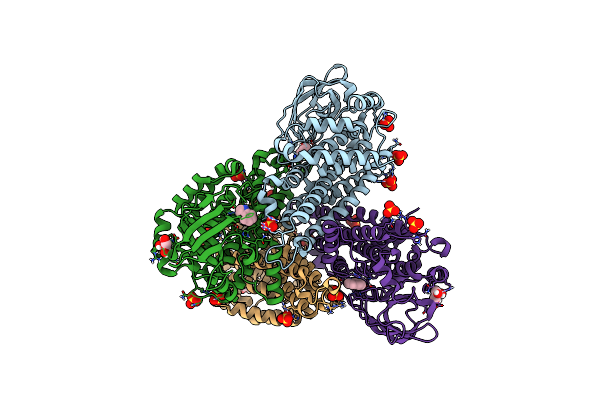 |
Organism: Paenibacillus sp. a11-2
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2017-08-09 Classification: OXIDOREDUCTASE Ligands: IND, SO4, GOL |
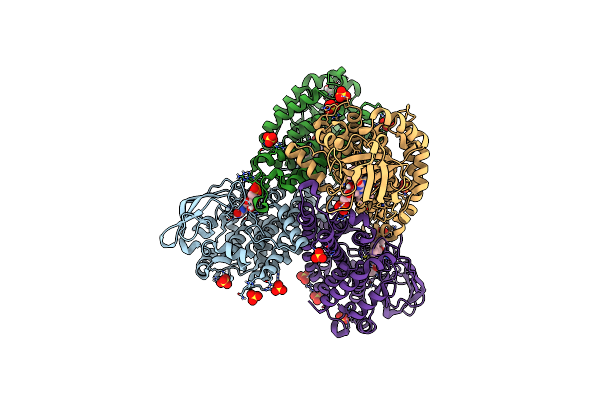 |
Crystal Structure Of Tertiary Complex Of Tdsc From Paenibacillus Sp. A11-2 With Fmn And Indole
Organism: Paenibacillus sp. a11-2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-08-09 Classification: OXIDOREDUCTASE Ligands: FMN, SO4, GOL, IND |
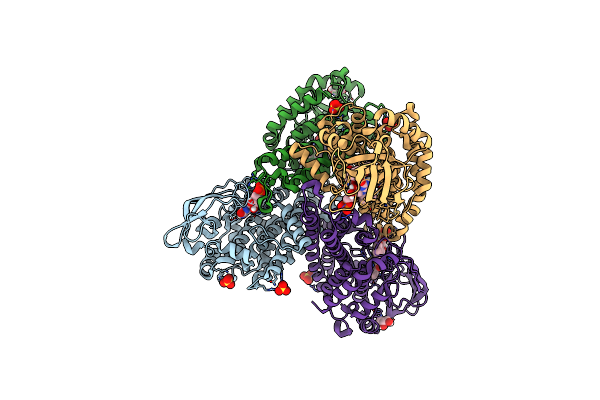 |
Crystal Structure Of Tertiary Complex Of Tdsc From Paenibacillus Sp. A11-2 With Fmn And Dibenzothiophene
Organism: Paenibacillus sp. a11-2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-08-09 Classification: OXIDOREDUCTASE Ligands: FMN, SO4, GOL, 83R |
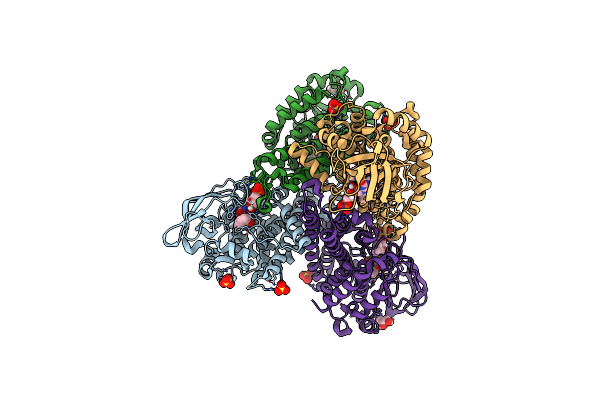 |
Crystal Structure Of Tertiary Complex Of Tdsc From Paenibacillus Sp. A11-2 With Fmn And Dibenzothiophene Sulfoxide
Organism: Paenibacillus sp. a11-2
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-08-09 Classification: OXIDOREDUCTASE Ligands: FMN, SO4, 83U, GOL |
 |
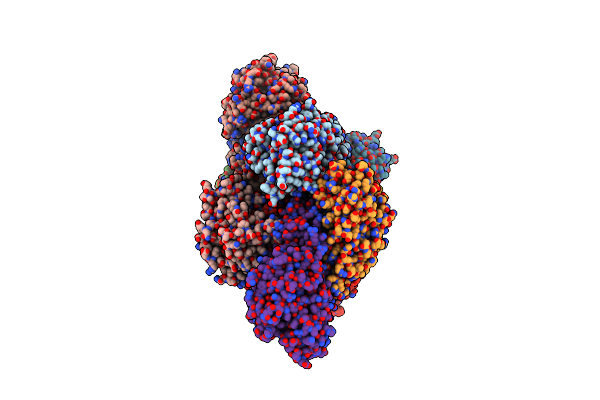
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-05-03 Classification: OXIDOREDUCTASE |
 |
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2015-02-25 Classification: OXIDOREDUCTASE |
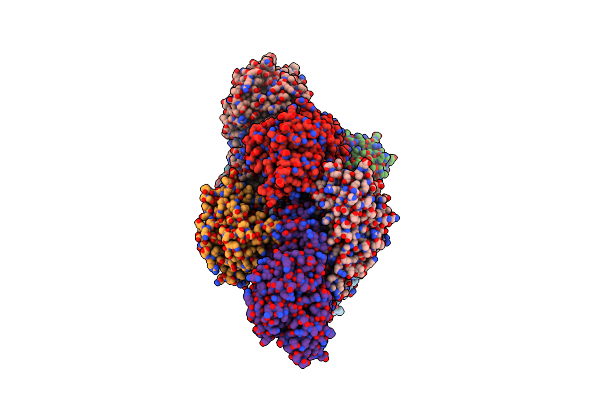 |
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-02-25 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-08-01 Classification: HYDROLASE Ligands: ACT, GOL |
 |
Crystal Structure Of Dszb C27S Mutant In Complex With 2'-Hydroxybiphenyl-2-Sulfinic Acid
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2006-08-01 Classification: HYDROLASE Ligands: ACT, OBP, MG |
 |
Crystal Structure Of Dszb C27S Mutant In Complex With Biphenyl-2-Sulfinic Acid
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-08-01 Classification: HYDROLASE Ligands: ACT, BPS |
 |
Recombinant Vanadium-Dependent Bromoperoxidase From Red Algae Corallina Pilulifera
Organism: Corallina pilulifera
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2003-09-30 Classification: HALOPEROXIDASE Ligands: PO4, CA |

