Search Count: 43
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.26 Å Release Date: 2019-03-20 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.23 Å Release Date: 2019-03-20 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-08-29 Classification: TRANSFERASE/IMMUNE SYSTEM Ligands: IRE, SO4 |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-08-29 Classification: TRANSFERASE/IMMUNE SYSTEM Ligands: IRE |
 |
Crystal Structure Of Hck Kinase Complexed With A Pyrrolo-Pyrimidine Inhibitor 7-[Trans-4-(4-Methylpiperazin-1-Yl)Cyclohexyl]-5-(4-Phenoxyphenyl)-7H-Pyrrolo[2,3-D]Pyrimidin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-06-06 Classification: TRANSFERASE Ligands: VSE |
 |
Parallel Homodimer Structures Of Voltage-Gated Sodium Channel Beta4 For Cell-Cell Adhesion
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-07-05 Classification: CELL ADHESION Ligands: P33, PG4, GOL |
 |
Parallel Homodimer Structures Of The Extracellular Domains Of The Voltage-Gated Sodium Channel Beta4 Subunit Explain Its Role In Cell-Cell Adhesion
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-07-05 Classification: CELL ADHESION Ligands: GOL |
 |
Crystal Structure Of Human Claudin-4 In Complex With C-Terminal Fragment Of Clostridium Perfringens Enterotoxin
Organism: Enterobacteria phage t4, Homo sapiens, Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2016-10-05 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of The Light-Driven Chloride Ion-Pumping Rhodopsin, Clp, From Nonlabens Marinus
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2016-07-13 Classification: TRANSPORT PROTEIN Ligands: RET, CL, HEX, OCT, D12, D10 |
 |
Structure-Based Site-Directed Photo-Crosslinking Analyses Of Multimeric Cell-Adhesive Interactions Of Vgsc Beta Subunits
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-06-08 Classification: METAL TRANSPORT |
 |
Organism: Cucumis melo
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-03-09 Classification: HYDROLASE Ligands: CL, PEG, GOL |
 |
Crystal Structure Of The Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, At 1.57 Angstrom
Organism: Acetabularia acetabulum
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2015-08-26 Classification: PROTON TRANSPORT Ligands: RET, OLB, D12, D10, OCT, C14 |
 |
Crystal Structure Of The Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, At 1.52 Angstrom
Organism: Acetabularia acetabulum
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2015-08-26 Classification: PROTON TRANSPORT Ligands: RET, OLB, D12, C14, R16, D10 |
 |
Crystal Structure Of The Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, At 1.80 Angstrom
Organism: Acetabularia acetabulum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-08-26 Classification: PROTON TRANSPORT Ligands: RET, OLB, C14, D10, R16, D12 |
 |
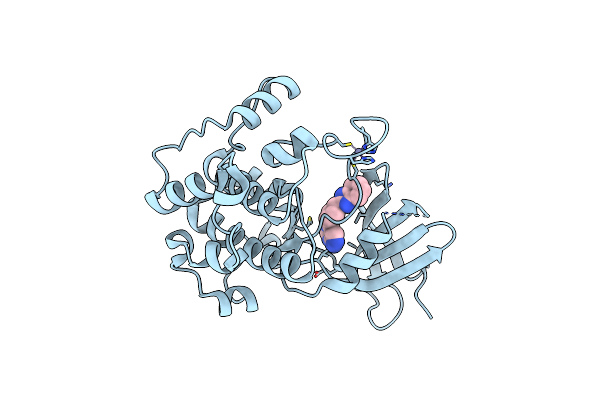
Crystal Structure Of S6K1 Kinase Domain In Complex With A Pyrimidine Derivative Pf-4708671 2-{[4-(5-Ethylpyrimidin-4-Yl)Piperazin-1-Yl]Methyl}-5-(Trifluoromethyl)-1H-Benzimidazole
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-08-06 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZN, 5FI |
 |
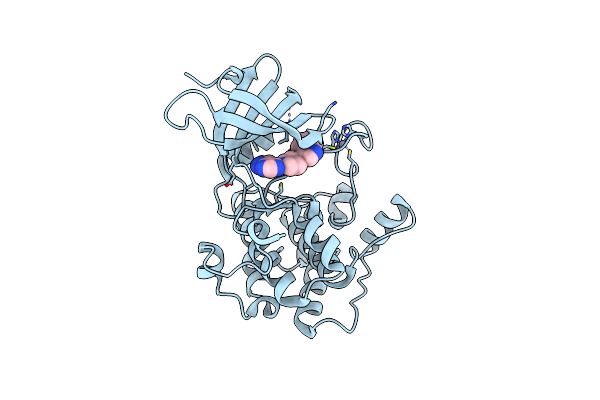
Crystal Structure Of S6K1 Kinase Domain In Complex With A Pyrazolopyrimidine Derivative 4-[4-(1H-Benzimidazol-2-Yl)Piperidin-1-Yl]-1H-Pyrazolo[3,4-D]Pyrimidine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-08-06 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZN, FZ8 |
 |
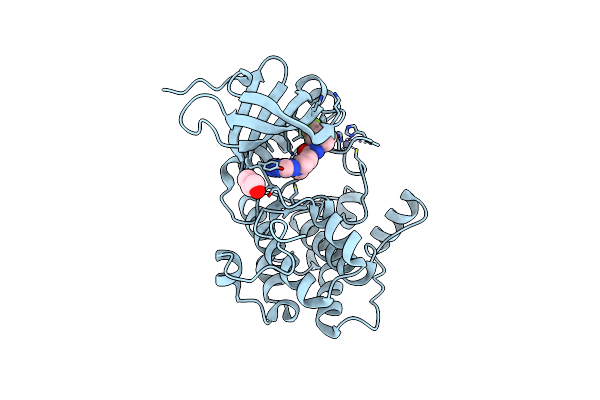
Crystal Structure Of S6K1 Kinase Domain In Complex With A Pyrazolopyrimidine Derivative 4-[4-(1H-Indol-3-Yl)-3,6-Dihydropyridin-1(2H)-Yl]-1H-Pyrazolo[3,4-D]Pyrimidine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2014-08-06 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZN, FZ9 |
 |
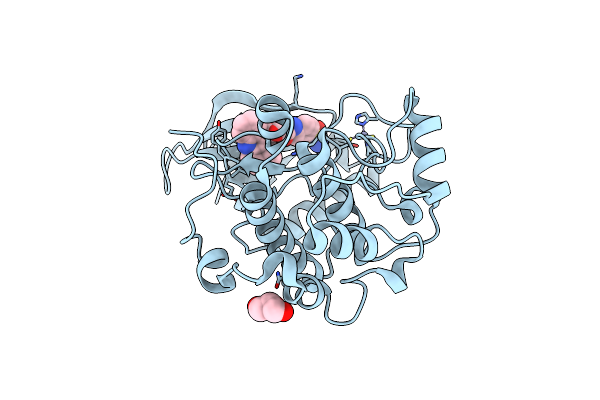
Crystal Structure Of S6K1 Kinase Domain In Complex With A Purine Derivative 1-(9H-Purin-6-Yl)-N-[3-(Trifluoromethyl)Phenyl]Piperidine-4-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-08-06 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZN, FS9, GOL |
 |
Crystal Structure Of S6K1 Kinase Domain In Complex With A Quinoline Derivative 2-Oxo-2-[(4-Sulfamoylphenyl)Amino]Ethyl 7,8,9,10-Tetrahydro-6H-Cyclohepta[B]Quinoline-11-Carboxylate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2014-08-06 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZN, F76, GOL |
 |
Crystal Structure Of S6K1 Kinase Domain In Complex With A Quinoline Derivative 1-Oxo-1-[(4-Sulfamoylphenyl)Amino]Propan-2-Yl-2-Methyl-1,2,3,4-Tetrahydroacridine-9-Carboxylate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2014-08-06 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZN, FS7, GOL |

