Search Count: 14
 |
Crystal Structure Of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed With Dinotefuran
Organism: Lymnaea stagnalis
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2024-12-18 Classification: SIGNALING PROTEIN Ligands: A1LW0, NAG |
 |
Crystal Structure Of Spiral2 Microtubule-Binding Domain From Physcomitrella Patens
Organism: Physcomitrium patens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-12-18 Classification: PLANT PROTEIN |
 |
Organism: Physcomitrium patens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-01-18 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of Cyclic Alpha-Maltosyl-1,6-Maltose Binding Protein From Arthrobacter Globiformis
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2020-12-02 Classification: SUGAR BINDING PROTEIN |
 |
Cyclic Alpha-Maltosyl-(1-->6)-Maltose Hydrolase From Arthrobacter Globiformis, Ligand-Free Form
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-09-12 Classification: HYDROLASE Ligands: CA |
 |
Cyclic Alpha-Maltosyl-(1-->6)-Maltose Hydrolase From Arthrobacter Globiformis, Complex With Cyclic Alpha-Maltosyl-(1-->6)-Maltose
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-09-12 Classification: HYDROLASE Ligands: CA |
 |
Cyclic Alpha-Maltosyl-(1-->6)-Maltose Hydrolase From Arthrobacter Globiformis, Complex With Panose
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2018-09-12 Classification: HYDROLASE Ligands: CA |
 |
Cyclic Alpha-Maltosyl-(1-->6)-Maltose Hydrolase From Arthrobacter Globiformis, Complex With Maltose
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-09-12 Classification: HYDROLASE |
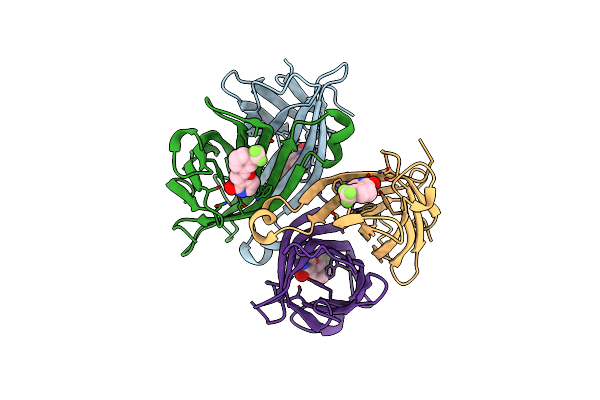 |
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2015-09-23 Classification: BIOTIN-Binding protein Ligands: T21 |
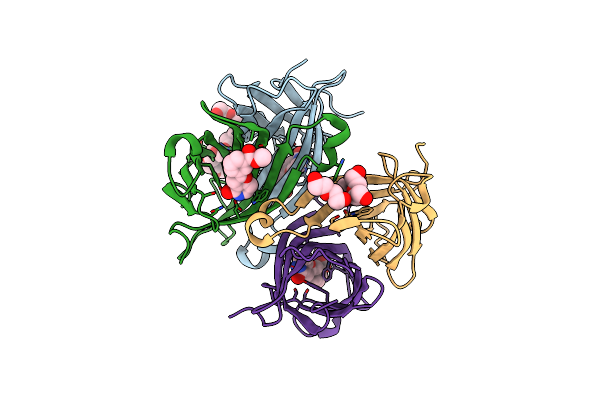 |
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2015-09-23 Classification: BIOTIN-BINDING PROTEIN Ligands: MT6, PE3, DMS, P6G |
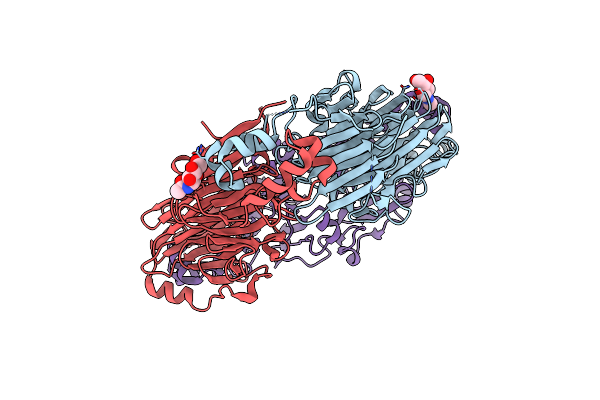 |
Crystal Structures Of Recombinant And Native Soybean Beta-Conglycinin Beta Homotrimers Complexes With N-Acetyl-D-Glucosamine
Organism: Glycine max
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2002-05-16 Classification: SUGAR BINDING PROTEIN Ligands: NAG |
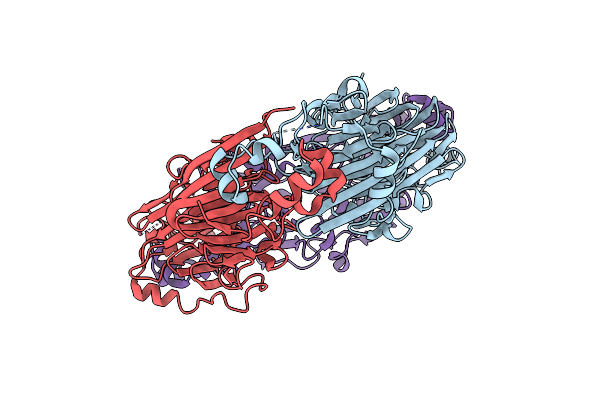 |
Crystal Structures Of Recombinant And Native Soybean Beta-Conglycinin Beta Homotrimers
Organism: Glycine max
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2002-05-16 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-09-13 Classification: NUCLEAR PROTEIN |
 |
Organism: Rhizopus niveus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1996-12-23 Classification: HYDROLASE (CARBOXYLIC ESTER) |

