Search Count: 17
 |
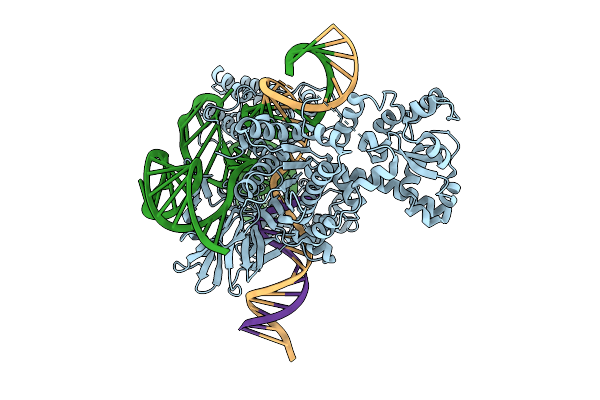
Cryo-Em Structure Of Esacas9_Nng-Guide Rna-Target Dna Complex In An Interrogation State
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN |
 |
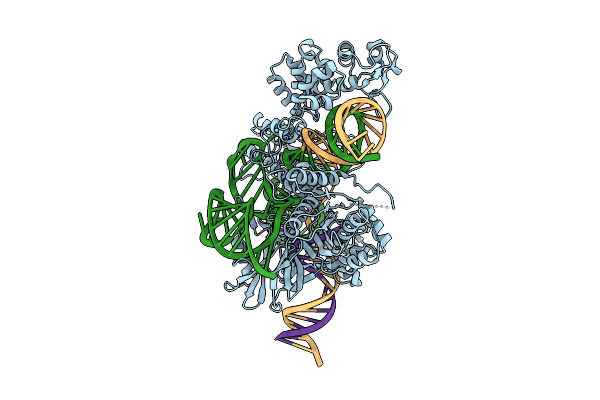
Cryo-Em Structure Of Esacas9_Nng-Guide Rna-Target Dna Complex In An Interrogation State
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN |
 |
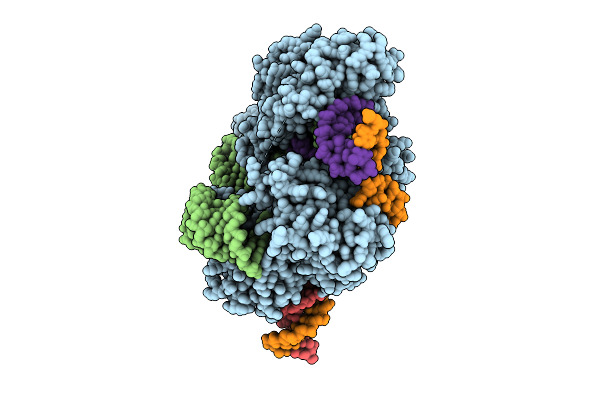
Cryo-Em Structure Of Esacas9_Nng-Guide Rna-Target Dna Complex In A Translocation State
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
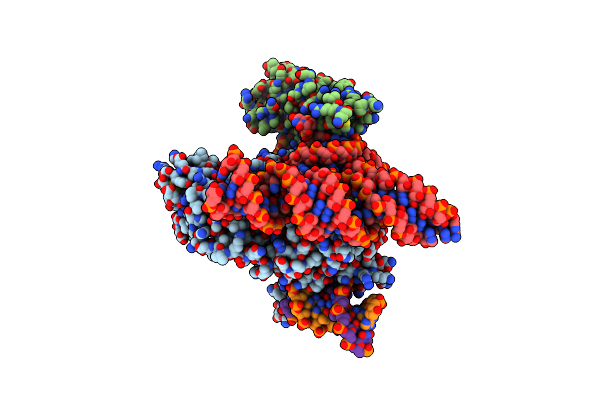
Cryo-Em Structure Of Esacas9_Nng-Guide Rna-Target Dna Complex In A Catalytically Active State
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Organism: Saccharolobus solfataricus
Method: X-RAY DIFFRACTION Release Date: 2024-08-07 Classification: OXIDOREDUCTASE Ligands: NAD, 69O |
 |
Organism: Saccharolobus solfataricus
Method: X-RAY DIFFRACTION Release Date: 2024-08-07 Classification: OXIDOREDUCTASE Ligands: AKG, NAD, EDO |
 |
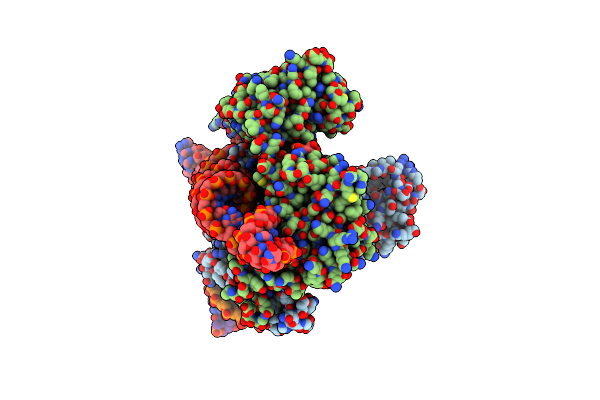
Organism: Acidibacillus sulfuroxidans, Acidibacillus sulfooxidans
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG, ZN |
 |
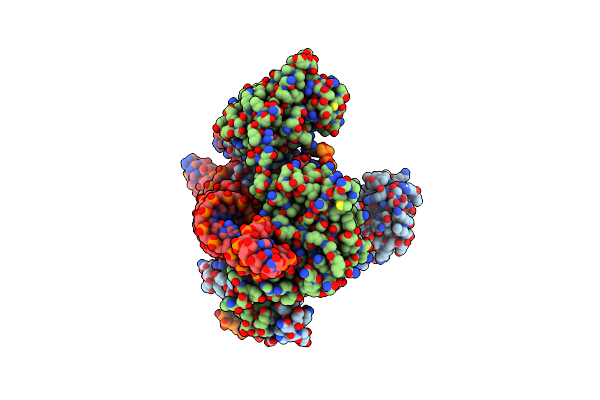
Organism: Sulfoacidibacillus thermotolerans
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: ZN, MG |
 |
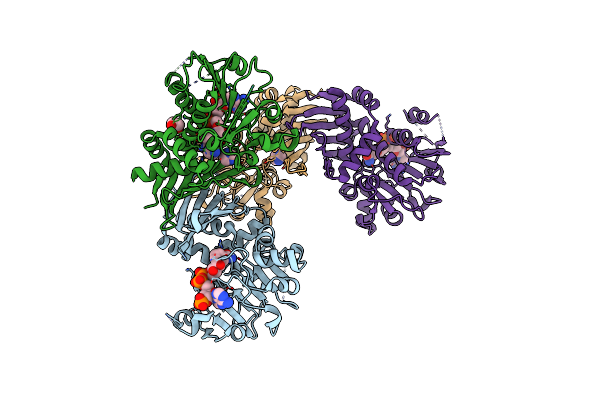
Organism: Sulfoacidibacillus thermotolerans
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: ZN, MG |
 |
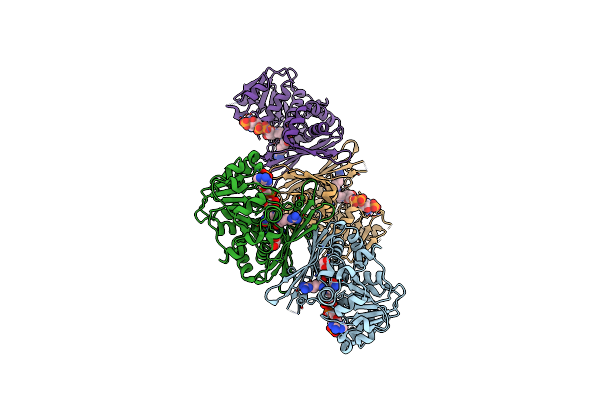
Organism: Pseudomonas veronii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-08-16 Classification: OXIDOREDUCTASE Ligands: NAP, LYS, IMD, EDO |
 |
Organism: Pseudomonas veronii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-08-16 Classification: OXIDOREDUCTASE Ligands: NDP, ARG, EDO |
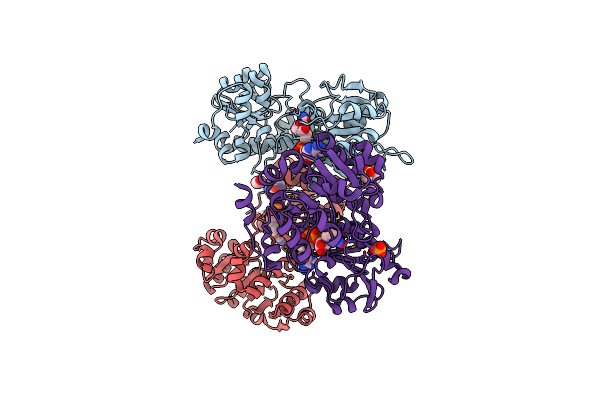 |
Organism: Geobacillus kaustophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-04-05 Classification: OXIDOREDUCTASE Ligands: NAD, EDO, EPE, PO4 |
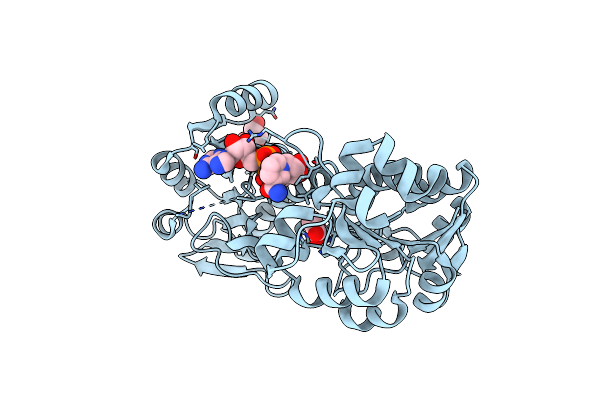 |
Organism: Geobacillus kaustophilus
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-04-05 Classification: OXIDOREDUCTASE Ligands: NAD, EDO, PYR |
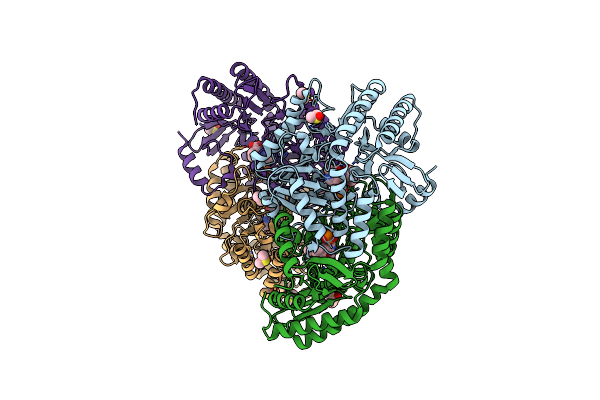 |
Organism: Lactobacillus buchneri
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2017-04-19 Classification: ISOMERASE Ligands: 7VO, DMS |
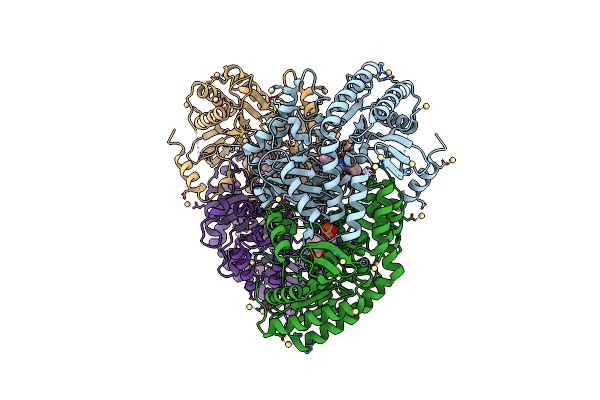 |
Organism: Lactobacillus buchneri
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2017-04-19 Classification: ISOMERASE Ligands: ILP, CD |
 |
Organism: Lactobacillus buchneri
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2016-04-20 Classification: ISOMERASE Ligands: PLP |
 |
Organism: Lactobacillus buchneri
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2016-04-20 Classification: ISOMERASE |

